New with 1.4.0
Starting with pandas 1.4.0, read_csv() delivers capability that allows you to handle these situations in a more graceful and intelligent fashion by allowing a callable to be assigned to on_bad_lines=.
For example, assume a CSV that could cause a bad data error: Expected 4 fields in line 3, saw 5:
C1,C2,C3,C4
10,11,12,13
25,26,27,28,garbage
80,81,82,83
The following lambda function simply ignores the last column in the bad line (as was desired in the original problem statement above):
df = pd.read_csv('your.csv', on_bad_lines=lambda x: x[:-1], engine='python')
df
C1 C2 C3 C4
0 10 11 12 13
1 25 26 27 28
2 80 81 82 83
The on_bad_lines callable function is called on each bad line and has a function signature (bad_line: list[str]) -> list[str] | None. If the function returns None, the bad line will be ignored. As you can see engine='python' is required.
The great thing about this is that it opens the door big-time for whatever fine-grained logic you want to code to fix the problem.
For example, say you’d like to remove bad data from the start or the end of the line and simply ignore the line if there is bad data in both the start and the end you could:
CSV
C1,C2,C3,C4
10,11,12,13
20,21,22,23,garbage
60,61,62,63
trash,80,81,82,83
trash,90,91,82,garbage
Function Definition
def line_fixer(x):
if not x[0].isnumeric() and x[-1].isnumeric():
return x[1:]
if not x[-1].isnumeric() and x[0].isnumeric():
return x[:-1]
return None
Result
df = pd.read_csv('your.csv', on_bad_lines=line_fixer, engine='python')
df
C1 C2 C3 C4
0 10 11 12 13
1 20 21 22 23
2 60 61 62 63
3 80 81 82 83
Содержание
- How to Drop Bad Lines with read_csv in Pandas
- How to Solve Error Tokenizing Data on read_csv in Pandas
- Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
- Step 2: Use correct separator to solve Pandas tokenizing error
- Step 3: Use different engine for read_csv()
- Step 4: Identify the headers to solve Error Tokenizing Data
- Step 5: Autodetect skiprows for read_csv()
- Conclusion
- How to Solve Python Pandas Error Tokenizing Data Error?
- Cause of the Problem
- Finding the Problematic Line (Optional)
- Using Err_Bad_Lines Parameter
- Using Python Engine
- Using Proper Separator
- Using Line Terminator
- Using header=None
- Using Skiprows
- Reading As Lines and Separating
- Conclusion
How to Drop Bad Lines with read_csv in Pandas
Here are two approaches to drop bad lines with read_csv in Pandas:
(1) Parameter on_bad_lines=’skip’ — Pandas >= 1.3
(2) error_bad_lines=False — Pandas ;
Date;Company A;Company A;Company B;Company B
2021-09-06;1;7.9;2;6
2021-09-07;1;8.5;2;7
2021-09-08;2;8;1;8.1
2021-09-09;2;8;1;»8.3;5.5″
Date;;Company A;;Company A;;Company B;;Company B
2021-09-06;;1;;7.9;;2;;6
2021-09-07;;1;;8.5;;2;;7
2021-09-08;;2;;8;;1;;8.1
2021-09-09;;2;;8;;1;;»8.3;;5.5″
Since Pandas 1.3 parameters error_bad_lines and warn_bad_linesbool will be deprecated.
Instead of them there will be new separator:
Specifies what to do upon encountering a bad line (a line with too many fields). Allowed values are :
- error , raise an Exception when a bad line is encountered.
- warn , raise a warning when a bad line is encountered and skip that line.
- skip , skip bad lines without raising or warning when they are encountered.
Note that depending on the separator:
The read_csv behavior can be different. You can check this article for more information: How to Use Multiple Char Separator in read_csv in Pandas
The reason for this is described in the documentation of Pandas:
Note that regex delimiters are prone to ignoring quoted data. Regex example: ‘rt’.
For example for a single separator «;» this code will work fine:
| Date | Company A | Company A.1 | Company B | Company B.1 |
|---|---|---|---|---|
| 2021-09-06 | 1 | 7.9 | 2 | 6 |
| 2021-09-07 | 1 | 8.5 | 2 | 7 |
| 2021-09-08 | 2 | 8.0 | 1 | 8.1 |
| 2021-09-09 | 2 | 8.0 | 1 | 8.3;5.5 |
While if you have a file with two separators you will get error or warning (depending on on_bad_lines ) for the quoted line:
Skipping line 5: Expected 5 fields in line 5, saw 6. Error could possibly be due to quotes being ignored when a multi-char delimiter is used.
In this case only 3 rows will be read from the CSV file
Источник
How to Solve Error Tokenizing Data on read_csv in Pandas
In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
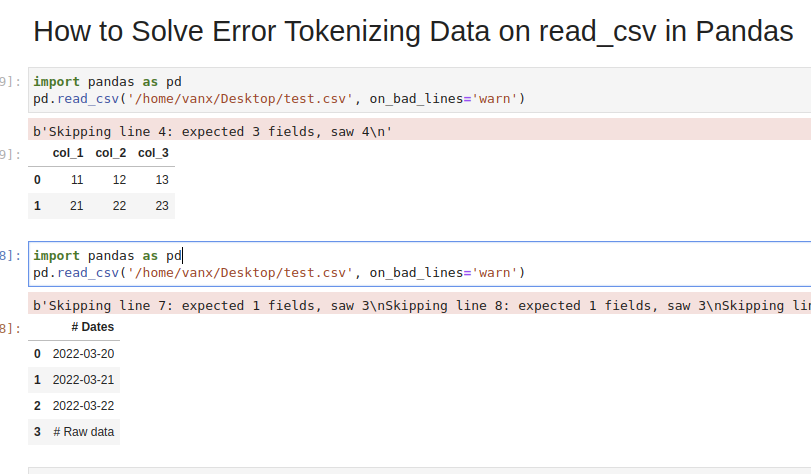
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
which we are going to read by — read_csv() method:
We will get an error:
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines=’skip’ in order to skip bad lines:
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
Note that delimiter is an alias for sep .
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
or skipping the last lines:
Note that we need to use python as the read_csv() engine for parameter — skipfooter .
In some cases header=None can help in order to solve the error:
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
Conclusion
In this post we saw how to investigate the error:
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
Источник
How to Solve Python Pandas Error Tokenizing Data Error?
While reading a CSV file, you may get the “Pandas Error Tokenizing Data“. This mostly occurs due to the incorrect data in the CSV file.
You can solve python pandas error tokenizing data error by ignoring the offending lines using error_bad_lines=False .
In this tutorial, you’ll learn the cause and how to solve the error tokenizing data error.
If you’re in Hurry
You can use the below code snippet to solve the tokenizing error. You can solve the error by ignoring the offending lines and suppressing errors.
Snippet
If You Want to Understand Details, Read on…
In this tutorial, you’ll learn the causes for the exception “Error Tokenizing Data” and how it can be solved.
Table of Contents
Cause of the Problem
- CSV file has two header lines
- Different separator is used
- r – is a new line character and it is present in column names which makes subsequent column names to be read as next line
- Lines of the CSV files have inconsistent number of columns
In the case of invalid rows which has an inconsistent number of columns, you’ll see an error as Expected 1 field in line 12, saw m . This means it expected only 1 field in the CSV file but it saw 12 values after tokenizing it. Hence, it doesn’t know how the tokenized values need to be handled. You can solve the errors by using one of the options below.
Finding the Problematic Line (Optional)
If you want to identify the line which is creating the problem while reading, you can use the below code snippet.
It uses the CSV reader. hence it is similar to the read_csv() method.
Snippet
Using Err_Bad_Lines Parameter
When there is insufficient data in any of the rows, the tokenizing error will occur.
You can skip such invalid rows by using the err_bad_line parameter within the read_csv() method.
This parameter controls what needs to be done when a bad line occurs in the file being read.
- False – Errors will be suppressed for Invalid lines
- True – Errors will be thrown for invalid lines
Use the below snippet to read the CSV file and ignore the invalid lines. Only a warning will be shown with the line number when there is an invalid lie found.
Snippet
In this case, the offending lines will be skipped and only the valid lines will be read from CSV and a dataframe will be created.
Using Python Engine
There are two engines supported in reading a CSV file. C engine and Python Engine.
C Engine
- Faster
- Uses C language to parse the CSV file
- Supports float_precision
- Cannot automatically detect the separator
- Doesn’t support skipping footer
Python Engine
- Slower when compared to C engine but its feature complete
- Uses Python language to parse the CSV file
- Doesn’t support float_precision . Not required with Python
- Can automatically detect the separator
- Supports skipping footer
Using the python engine can solve the problems faced while parsing the files.
For example, When you try to parse large CSV files, you may face the Error tokenizing data. c error out of memory. Using the python engine can solve the memory issues while parsing such big CSV files using the read_csv() method.
Use the below snippet to use the Python engine for reading the CSV file.
Snippet
This is how you can use the python engine to parse the CSV file.
Optionally, this could also solve the error Error tokenizing data. c error out of memory when parsing the big CSV files.
Using Proper Separator
CSV files can have different separators such as tab separator or any other special character such as ; . In this case, an error will be thrown when reading the CSV file, if the default C engine is used.
You can parse the file successfully by specifying the separator explicitly using the sep parameter.
As an alternative, you can also use the python engine which will automatically detect the separator and parse the file accordingly.
Snippet
This is how you can specify the separator explicitly which can solve the tokenizing errors while reading the CSV files.
Using Line Terminator
CSV file can contain r carriage return for separating the lines instead of the line separator n .
In this case, you’ll face CParserError: Error tokenizing data. C error: Buffer overflow caught — possible malformed input file when the line contains the r instead on n .
You can solve this error by using the line terminator explicitly using the lineterminator parameter.
Snippet
This is how you can use the line terminator to parse the files with the terminator r .
CSV files can have incomplete headers which can cause tokenizing errors while parsing the file.
You can use header=None to ignore the first line headers while reading the CSV files.
This will parse the CSV file without headers and create a data frame. You can also add headers to column names by adding columns attribute to the read_csv() method.
Snippet
This is how you can ignore the headers which are incomplete and cause problems while reading the file.
Using Skiprows
CSV files can have headers in more than one row. This can happen when data is grouped into different sections and each group is having a name and has columns in each section.
In this case, you can ignore such rows by using the skiprows parameter. You can pass the number of rows to be skipped and the data will be read after skipping those number of rows.
Use the below snippet to skip the first two rows while reading the CSV file.
Snippet
This is how you can skip or ignore the erroneous headers while reading the CSV file.
Reading As Lines and Separating
In a CSV file, you may have a different number of columns in each row. This can occur when some of the columns in the row are considered optional. You may need to parse such files without any problems during tokenizing.
In this case, you can read the file as lines and separate it later using the delimiter and create a dataframe out of it. This is helpful when you have varying lengths of rows.
In the below example,
- the file is read as lines by specifying the separator as a new line using sep=’n’ . Now the file will be tokenized on each new line, and a single column will be available in the dataframe.
- You can split the lines using the separator or regex and create different columns out of it.
- expand=True expands the split string into multiple columns.
Use the below snippet to read the file as lines and separate it using the separator.
Snippet
This is how you can read the file as lines and later separate it to avoid problems while parsing the lines with an inconsistent number of columns.
Conclusion
To summarize, you’ve learned the causes of the Python Pandas Error tokenizing data and the different methods to solve it in different scenarios.
Different Errors while tokenizing data are,
- Error tokenizing data. C error: Buffer overflow caught — possible malformed input file
- ParserError: Expected n fields in line x, saw m
- Error tokenizing data. c error out of memory
Also learned the different engines available in the read_csv() method to parse the CSV file and the advantages and disadvantages of it.
You’ve also learned when to use the different methods appropriately.
Источник
In this tutorial, we’ll show how to use read_csv pandas to import data into Python, with practical examples.
csv (comma-separated values) files are popular to store and transfer data. And pandas is the most popular Python package for data analysis/manipulation. These make pandas read_csv a critical first step to start many data science projects with Python.
You’ll learn from basics to advanced of pandas read_csv, how to:
- import csv files to pandas DataFrame.
- specify data types (low_memory/dtype/converters).
- use a subset of columns/rows.
- assign column names with no header.
- And More!
This pandas tutorial includes all common cases when loading data using pandas read_csv.
Practice along and keep this as a cheat sheet as well!
Table Of Contents
- Before reading the files
- Pandas Data Structures: Series? DataFrame?
- pandas read_csv Basics
- Fix error_bad_lines of more commas
- Specify Data Types: Numeric or String
- Specify Data Types: Datetime
- Use certain Columns (usecols)
- Set Column Names (names/prefix/no header)
- Specify Rows/Random Sampling (nrows/skiprows)
- pandas read_csv in chunks (chunksize) with summary statistics
- Load zip File (compression)
- Set Missing Values Strings (na_values)
- pandas read_csv Cheat Sheet
Before reading the files
If you have no experience with Python, please take our FREE Python crash course: breaking into data science.
Learn Python for data science: FREE online course – Just into Data
A FREE Python online course, beginner-friendly tutorial. Start your successful data science career journey: learn Python for data science, machine learning.
To make it easy to show, we created 5 small csv files. You can download them to your computer from Github pandas-read-csv-practice to practice.
Note: You can open these csv files and view them through Jupyter Notebook. Just launch a Jupyter Notebook, then look for them within the directory.
Or for Windows users, you can right-click the file, select “Edit” and view it in Notepad. For Mac users, you can right-click and open with TextEdit.
The comma (,) is used to separate columns, while a new line is used to separate rows.
To start practicing, you can either:
- open the read_csv_code.ipynb file from Jupyter Notebook.
or - create a new notebook within the same folder as these csv files.
Note: To avoid specifying the whole path/directory for the folder, Python needs both the csv files and the notebook to be in the same directory. This way, Python can locate the files by its name in the current working directory.
Otherwise, you will get FileNotFoundError.
To be able to use the pandas read_csv function, we also need to import the pandas and NumPy packages into Python. Both libraries should have been installed on your computer, if you installed Anaconda Distribution.
Note: pd is the common alias name for pandas, and np is the common alias name for NumPy.
Pandas Data Structures: Series? DataFrame?
pandas is the most popular Python data analysis/manipulation package, which is built upon NumPy.
pandas has two main data structures: Series and DataFrame.
- Series: a 1-dimensional labeled array that can hold any data type such as integers, strings, floating points, Python objects.
It has row/axis labels as the index. - DataFrame: a 2-dimensional labeled data structure with columns of potentially different types.
It also contains row labels as the index.
DataFrame can be considered as a collection of Series; it has a structure like a spreadsheet.
We’ll be using the read_csv function to load csv files into Python as pandas DataFrames.
Great!
With all this basic knowledge, we can start practicing pandas read_csv!
pandas read_csv Basics
There is a long list of input parameters for the read_csv function. We’ll only be showing the popular ones in this tutorial.
The most basic syntax of read_csv is below.
With only the file specified, the read_csv assumes:
- the delimiter is commas (,) in the file.
We can change it by using the sep parameter if it’s not a comma. For example, df = pd.read_csv(‘test1.csv’, sep= ‘;’) - the first row of the file is the headers/column names.
- read all the data.
- the quote character is double (“).
- an error will occur if there are bad lines.
Bad lines happen when there are too many delimiters in the row.
Most structured datasets that are saved to text-based files can be opened using this method. They often have clear formats and don’t need any further specification of read_csv.
The test1.csv file is nice and clean, so the default setting is appropriate to load the file.
Below you can see the original file (left) and the pandas DataFrame df (right).
Note: you can use the type function to find out that df is a pandas.core.frame.DataFrame.
As you can see, the first row in the csv file was taken as the header, and the first three lines are straightforward.
But how can we have commas (,) and double quotes (“) in the 3rd and 4th row?
Shouldn’t they be special characters?
Take a closer look at the original file of test1.csv, and you will notice the tricks:
- to have the delimiter comma (,) in the data, we need to put quotes around them.
- to have the quote (“) in the data, we need to type two double quotes (“”) to help read_csv understand that we want to read it literally.
Fix error_bad_lines of more commas
The most common error is when there are extra delimiter characters in a row.
This usually happens when the comma in the data wasn’t quoted, which often appears for variables of addresses or company names.
Let’s see an example.
Below is the original data for test2.csv. We can see that among the row for CityD, there’s an extra comma (,) within the address (58 Fourth Street, Apt 500). But the entry isn’t quoted.
If we read by using the default settings, read_csv will be confused by this extra delimiter and give an error.
As the error message says, Python expected 3 fields in line 5, but saw 4. This is due to the extra comma.
There are two main methods to fix this error:
- the best way is to correct the error within the original csv file.
- when not possible, we can also skip the bad lines by changing the error_bad_lines parameter setting to be False.
This will load the data into Python while skipping the bad lines, but with warnings.
b'Skipping line 5: expected 3 fields, saw 4n'
We can also suppress this warning by setting warn_bad_lines=False, but we’d like to see the warnings most of the time.
As you can see, the line with CityD was skipped here.
Next, let’s see another common issue with csv files.
Specify Data Types: Numeric or String
As you know, the csv files are plain-text files.
While it is important to specify the data types such as numeric or string in Python. We need to rely on pandas read_csv to determine the data types.
By default, if everything in a column is number, read_csv will detect that it is a numerical column; if there are any non-numbers in the column, read_csv will set the column to be an object type.
It is more apparent with an example.
The test3.csv has three columns as below. The address column has both numbers and strings.
If we use the default settings of read_csv to load its data.
read_csv specifies the data types of:
- id as int64 (integer) since it contains only numbers.
- address as object, since it contains some text, even though most of the lines are numbers.
- city as object since it’s all text.
This is great since these are the data types we want these columns to have.
The problem of Mixed Data Types
But when the file has many rows, we might get a mixed data type column. In this situation, there will be problems.
To show this, let’s create a large csv file with 2 columns and 1,000,010 rows:
- col1 has the letter ‘a’ in every row.
- col2 has 1,000,000 rows with the number 123 and the last 10 rows with the string ‘Hello’.
If we use read_csv with the default settings, pandas will get confused and gives a warning saying it is mixed data types.
/Users/justin/py_envs/DL/lib/python3.7/site-packages/IPython/core/interactiveshell.py:3063: DtypeWarning: Columns (1) have mixed types.Specify dtype option on import or set low_memory=False. interactivity=interactivity, compiler=compiler, result=result)
So even though Python loaded the data, it has a strange data type. Python considers the first 1,000,000 rows with 123 as integer/numeric, and the last 10 rows with ‘Hello’ as strings.
We can perform numeric operations on the first 20 rows of 123.
But we can’t perform any numerical or string operations on the entire column.
The first “+100” operation returns an error “TypeError: can only concatenate str (not “int”) to str”. While the second str.len() function returns NaN for all the 123 rows.
And we can take out two random rows to check their data types. We can see that the row 0 with 123 is an integer, while the row 1,000,009 with ‘Hello’ is a string.
This column has mixed data types. This is NOT what we want!
How do we fix them?
There are 3 main ways. Let’s look at them one-by-one.
Method #1: set low_memory = False
By default, low_memory is set to True. That means when the file is larger, read_csv loads the file in chunks.
If an entire chunk has all numeric values, read_csv will save it as numeric.
If it encounters some non-numeric values in a chunk, then it will save the values in that chunk as non-numeric (object).
This is why we had the mixed data type in our large file example.
When we set low_memory = False, read_csv reads in all rows together and decides how to store the values based on all the rows. If there are any non-numeric values, then it will store as an object.
Let’s try it out.
After it’s loaded, we can test the data types by taking out one row with 123 and one row with ‘Hello’ from the DataFrame df again.
It will tell us that Python is now treating them both as str (strings).
And we can perform string operations on the entire column. Try it out!
But using low_memory = False is not memory efficient, which leads to better method #2.
Method #2: set dtype (data types)
It is more efficient to tell Python the data types (dtype) when loading the data, especially when the dataset is larger.
Let’s try setting the column to be the string data type as below.
We’ll see that the string operation str.len() returns the result with no issue.
What if we know that this column should be numeric, and those text entries are typos?
Note that we cannot use a numeric type in this situation, because there are texts ‘Hello’ in the column.
But we can use method #3 to convert the values in the column.
Method #3: set converters (functions)
We can define a converter function to convert the values in specific columns.
In the example below, we kept the values that are numeric and changed the text values to all NaNs for col2.
Then we use this converter_func as a parameter in read_csv.
Numeric and strings are the most common data types, and we now know how to deal with them!
What about another common data type: date and time?
Specify Data Types: Datetime
When we have date/time columns, we cannot use the dtype parameter to specify the data type.
We’ll need to use the parse_dates, date_parser, dayfirst, keep_date_col parameters within the read_csv function.
We won’t go over it here, but please take a look at the official document for more details.
And if you do use date_parser, it is good to get familiar with the Python DateTime formats.
Further Reading: How to Manipulate Date And Time in Python Like a Boss
So far we’ve been loading the entire file, what if we only want a subset?
Use certain Columns (usecols)
When we only want to analyze certain columns from the file, it saves memory to only read in those columns.
We can use the usecols parameter.
For example, only the col2 is loaded with the specification below.
Since we are looking at columns, let’s also see how to name them better.
Set Column Names (names/prefix/no header)
When we don’t have a header row in the csv file, we can specify columns names.
For example, test5.csv is created without a header row.
If we use the default setting to read_csv, Python will take the first row as the column names.
To fix this, we can use the names parameter to set the column names as my_col1, mycol2, mycol3.
What if we have many columns, and we want to assign them names with the same format/prefix to them?
We can set header = None, and the prefix parameter. read_csv will assign column names with the prefix.
After looking at columns, let’s see how we can deal with rows with more flexibility.
Specify Rows/Random Sampling (nrows/skiprows)
When we have a large dataset, it might not all fit into memory. Rather than importing the entire dataset, we can take a subset/sample from it.
Method #1: use nrows
We can use the nrows parameter to set the top number of rows to read from the file.
For example, we can set nrows = 10 to read the first 10 rows.
Method #2: use skiprows
Using the skiprows parameter gives us more flexibility for the rows to skip.
But we do need to provide more information. We have to input:
- an integer showing the number of rows to skip at the start of the file.
- or a list of row numbers (starting at 0 index position) to skip.
- or a callable function that returns either True (skip) or False (otherwise) for each row number.
Let’s start with a simple example.
We can set skiprows=100,000 to skip the first 100,000 rows.
If we check the shape of the DataFrame, it will return (900010, 2), which is 100,000 rows less than the original file.
When we set skiprows as a list of numbers, read_csv will skip the row numbers in the list.
For example, to achieve random sampling, we can first create a random list skip_list of size 200,000 from a range(1,000,010). Then we can use skiprows to skip the 200,000 random rows specified by the skip_list.
In this case, we know the total number of rows in the file is 1,000,010, so we can draw random row numbers from range(1,000,010).
What if we don’t know that?
We can create a function that randomly samples some of the rows.
For example, the code below uses a lambda function to sample roughly 10% of the rows randomly. The lambda function goes through each row index, and there’s a 10% chance that a particular row is included in the new dataset.
Note that we also skipped the first row (x == 0) containing the header since we are using names to specify the column names.
pandas read_csv in chunks (chunksize) with summary statistics
When we have a really large dataset, another good practice is to use chunksize.
As mentioned earlier as well, pandas read_csv reads files in chunks by default. But it keeps all chunks in memory.
While with the chunksize setting, Python reads in chunks without keeping them in memory until it’s called. This is more efficient and makes it easier to spot any errors when loading the data.
Let’s see how it works.
For example, we use the chunksize setting to create a TextFileReader object reader below.
Note that reader is not a pandas DataFrame anymore. It is a pandas TextFileReader. The data is not in memory until we call it.
We can use the get_chunk method to fetch chunks from the file.
You may try out the below code to see what it returns.
A more popular way of using chunk is to loop through it and use aggregating functions of pandas groupby to get summary statistics.
For example, we can iterate through reader to process the file by chunks, grouping by col2, and counting the number of values within each group/chunk.
Related article: How to GroupBy with Python Pandas Like a Boss
If you are not familiar with pandas GroupBy, take a look at this complete tutorial with examples.
Based on the output above, we can do another GroupBy to return the total count for each group in col2.
Load zip File (compression)
read_csv also supports reading compressed files. This is very useful since we often store the csv file compressed to save storage space.
For example, we saved a zip version of test4.csv within the folder. And we can use the compression parameter setting to read it directly.
Set Missing Values Strings (na_values)
By default the following values are interpreted as NaN/missing value: ‘’, ‘#N/A’, ‘#N/A N/A’, ‘#NA’, ‘-1.#IND’, ‘-1.#QNAN’, ‘-NaN’, ‘-nan’, ‘1.#IND’, ‘1.#QNAN’, ‘<NA>’, ‘N/A’, ‘NA’, ‘NULL’, ‘NaN’, ‘n/a’, ‘nan’, ‘null’.
We can also specify extra strings within the file to be recognized as NA/NaN by using the na_values parameter.
pandas read_csv Cheat Sheet
The code below summarized everything covered in this tutorial. You’ve learned a lot of pandas read_csv parameters!
That’s it for this pandas tutorial for pandas read_csv. You’ve mastered this first step of your data science projects!
Leave a comment for any questions you may have or anything else.
Related “Break into Data Science” resources:
Python crash course: Break into Data Science – FREE
Learn Python for data science: FREE online course – Just into Data
A FREE Python online course, beginner-friendly tutorial. Start your successful data science career journey: learn Python for data science, machine learning.
How to GroupBy with Python Pandas Like a Boss
Read this pandas tutorial to learn Group by in pandas. It is an essential operation on datasets (DataFrame) when doing data manipulation or analysis.
How to Learn Data Science Online: ALL You Need to Know
Check out this for a detailed review of resources online, including courses, books, free tutorials, portfolios building, and more.
Of course @WillAyd, sorry for the delay ;
-
where the «implicit index» detection is defined and performed :
1) Look for implicit index: there are more columns -
where it is used to remove the error :
We have unexpected results here when specifying index_col=False because it disable altogether lines too long :
| self.index_col is not False and |
Synthesis of the outputs according to inputs :
names = ['ID', 'X1', 'X2', 'X3'] # With malformed line on line 3 s = """0,1,2,3 1,2,3,4 4,3,2,1,5""" pd.read_csv(io.StringIO(s), names=names, error_bad_lines=False, header=None, engine='python') # Output : Good ID X1 X2 X3 0 0 1 2 3 1 1 2 3 4 Skipping line 3: Expected 4 fields in line 3, saw 5 # With malformed line on line 2 s = """0,1,2,3 1,2,3,4,5 4,3,2,1""" pd.read_csv(io.StringIO(s), names=names, error_bad_lines=False, header=None, engine='python') # Output : Good or Bad ? ID X1 X2 X3 0 1 2 3 NaN 1 2 3 4 5.0 4 3 2 1 NaN # With index_col = False and malformed line on line 2 (or 3, it doesn't matter) s = """0,1,2,3 1,2,3,4,5 4,3,2,1""" pd.read_csv(io.StringIO(s), names=names, error_bad_lines=False, header=None, engine='python', index_col=False) # Output : Bad ID X1 X2 X3 0 0 1 2 3 1 1 2 3 4 2 4 3 2 1