Содержание
- Ошибка при установке пакета ‘графика’ (но не других) в R
- Error in install.packages : object ‘needed_packages’ not found #21
- Comments
- Troubles with Learn Lesson and bridging data into R
- Credibly Curious
- 2020-05-29
- Error in loadNamespace(Name) : There is No Package Called ‘here’
- Nicholas Tierney
- Install the package
- Why does that work?
- Another Error: Error in install.packages : object ‘here’ not found
- Another error: Warning in install.packages : package ‘emo’ is not available (for R version 3.6.2)
- Problem solving: Is the package perhaps misspelt?
- Problem solving: Does the package exist on CRAN?
- Why write this blog post?
- Thanks
Ошибка при установке пакета ‘графика’ (но не других) в R
Я новичок в R и новичок в StackOverflow, и я впервые задаю вопрос, надеюсь, я соблюдаю все правила .
В любом случае. Я установил R (версия 3.5.0) и R-Studio на моем ноутбуке с установленной Win 10 Home (в той же родительской папке ‘D: ’) и сбросил каталог временных файлов в папку в D: , чтобы китайский в C: Users 中文 documents (который является местом по умолчанию для файлов темпа) не влияет на соединение между R-studio и R (мне сказали, что любой соответствующий каталог должен быть на английском языке, чтобы R-Studio работала) .
Проблема в том, что я не могу установить пакет ‘graphics’ при запуске
Полученная ошибка говорит примерно так:
Это довольно странно, так как пакет ‘graphics’ довольно простой, и на самом деле я не нашел никого, кто сообщал бы о таких ошибках в Интернете.
Меня также беспокоит, что другие пакеты, такие как «прогноз» и «XML», могут быть успешно установлены без каких-либо отчетов об ошибках.
Я рассмотрел несколько похожих вопросов в StackOverflow, имея отчеты об ошибках с «Ошибка в gzfile (файл, «wb»): не удается открыть соединение», но у всех, похоже, есть что-то еще, чтобы завершить предложение. В моем случае он просто останавливает описание ошибки с помощью «не могу открыть соединение». Также я не мог найти людей, у которых были бы похожие проблемы с «Ошибка сохранения сеанса (параметры): ошибка выполнения кода R«.
Я понимаю, что это довольно необычно. Я даже не могу найти подходящие теги, кроме r для этого вопроса. Хотя кажется, что ни у кого нет проблем с «графикой», эта проблема меня действительно беспокоит, и я надеюсь, что кто-нибудь сможет предложить возможные решения. Большое спасибо.
Похоже, он у вас уже установлен. У вас ошибка при записи library(graphics)
гы . Он у меня уже есть . Но почему мне не нужно написать код на другом ноутбуке, чтобы использовать функцию «сюжет»? это потому, что я никогда не удалял файл R.history?
Я не уверен, что понимаю. У вас есть сеанс, который не распознает функцию plot ? Если да, то сначала попробуйте сделать library(graphics) ?
(даже не знаю, что означает сеанс, но) Да. Я извлек фрейм данных с веб-сайта (с функцией в пакете XML) и попытался построить один столбец, но он сообщил кое-что, чего я сейчас не помню. Чтобы убедиться, что я не злоупотребляю функцией «plot», я написал «? Plot», и в боковом поле не было никакой информации, которую я ожидал. Вместо этого было сказано что-то вроде того, что у меня этого не было. Вот почему я запустил install.packages (‘graphics’) ‘, потому что думал, что у меня его нет на только что установленном R
Хорошо, сначала попробуйте еще раз с library(graphics) . Я только предполагаю, что по какой-то причине этот пакет не загружается при запуске.
Большое спасибо. Эта проблема задержала меня на целый день. Несколько часов назад я очень боялся спросить о StackOverflow, потому что чувствовал, что что-то тупо не так.
Источник
Error in install.packages : object ‘needed_packages’ not found #21
After successfully updating R from 3.3. to 3.5 using updateR() from RStudio and selecting «yes» to restarting R, I got the following message:
It also looks like my old packages are no longer installed.
Here is my sessionInfo():
The text was updated successfully, but these errors were encountered:
I am having exactly the same problem. My sessionInfo() is identical to that listed above. The only difference is that I was upgrading from R version 3.4.3.
Everything went smoothly, R was updated to R version 3.5.0 (2018-04-23) — «Joy in Playing»
Also the packages installed on your previous version of R were restored
[At this point RStudio told me it was trying to update loaded packages and asked to restart, so I said yes]
Restarting R session.
> install.packages(as.vector(needed_packages))
Error in install.packages : object ‘needed_packages’ not found
R version change [3.4.3 -> 3.5.0] detected when restoring session; search path not restored
Here’s the list of packages that seem to be currently installed after running updateR() — it seems that the only ones remaining are those that ship with R («base» or «recommended» priority).
hi @prestongreene and @cring-toxstrategies , package restoring is one of the last features developed so thank you for the precious feedback. Going to work on this.
Источник
Troubles with Learn Lesson and bridging data into R
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I am attempting to complete the Learn Lesson Perform ecological niche factor analysis in R—Identify an Ecological Niche for African Buffalo but am having troubles with bridging my data into R. Specifically, step 6 onward: the following function exactly as it appears in step 6 of the lesson simply does not work for me: arc.raster(data_path) . When I attempt to enter this in RStudio, I keep getting the message Error in arc.raster(data_path) : object ‘data_path’ not found
I am running ArcGIS Pro 2.2.4 and RStudio Version 1.1.463 on Windows 7. I do not know a lot about using R in general, but I thought a learn lesson would be a good way to check it out. This may be a very simple thing to fix but I simply don’t know enough to troubleshoot it.
What could be the problem?
Thanks in advance!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I am sorry to hear that you are running into problems, hopefully I can help. Would you be able to send me what you entered in step 4 of the lesson and what the R console returned to you after entering that code? Step 4 asks that you define the variable ‘data_path’ by entering the following code:
Just as a note though, the location you saved your data at might be different. For example, you might have picked a different folder location and as a result, will need to adjust the path to reflect the folders you used to store the lesson’s data in. Additionally, R only recognizes forward slashes («/») and when you copy a path directly from Windows File Explorer, it contains back slashes («») so those need to be adjusted in order for R to know where your data is located.
I am curious what happened after this step, such as if R threw a warning or error. The reason I ask is because the current error you are seeing says that the variable ‘data_path’ is not defined. This means that R is says it unaware of any variable called ‘data_path’ and as a result, is unsure of how to handle your most recent line of code since it uses that variable.
Let me know when you have the chance. Thank you!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I figured out that I must have been making a silly mistake in identifying the variable ‘data_path’. I see what I was doing wrong there now.
However, I have run into a different problem: Step 1 under the section Produce a habitat suitability map. When I attempt to load the sp package, R asks me to restart because the package is already currently loaded (it apparently loaded when I used library(raster) earlier in the session). If I click Yes to restart R I end up in a loop where R keeps wanting to restart (and interestingly it also gives a Windows message that it has stopped working, though R stays open i.e. does not crash). If I click No or Cancel, I simply cannot load the sp package.
Below is a copy/paste of my RStudio console (at the end I attempted to let R restart twice by clicking Yes, then I clicked No).
> library(arcgisbinding)
*** Please call arc.check_product() to define a desktop license.
> arc.check_product()
product: ArcGIS Pro ( 12.2.0.12813 )
license: Advanced
version: 1.0.1.232
> data_path install.packages(‘raster’)
trying URL ‘ https://cran.rstudio.com/bin/windows/contrib/3.5/raster_2.8-4.zip ‘
Content type ‘application/zip’ length 3769843 bytes (3.6 MB)
downloaded 3.6 MB
package ‘raster’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:UserskflammAppDataLocalTempRtmpM9T3Padownloaded_packages
> library(raster)
Loading required package: sp
> arc_raster r_raster variables variables$pixel_index variables pixel_index variables$pixel_index ecological_variables buffalo_presence install.packages(«ade4»)
trying URL ‘ https://cran.rstudio.com/bin/windows/contrib/3.5/ade4_1.7-13.zip ‘
Content type ‘application/zip’ length 5201783 bytes (5.0 MB)
downloaded 5.0 MB
package ‘ade4’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:UserskflammAppDataLocalTempRtmpM9T3Padownloaded_packages
> library(ade4)
> dudi_obj install.packages(«adehabitatHS»)
trying URL ‘ https://cran.rstudio.com/bin/windows/contrib/3.5/adehabitatHS_0.3.13.zip ‘
Content type ‘application/zip’ length 1515084 bytes (1.4 MB)
downloaded 1.4 MB
package ‘adehabitatHS’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:UserskflammAppDataLocalTempRtmpM9T3Padownloaded_packages
> library(adehabitatHS)
Loading required package: adehabitatMA
Attaching package: ‘adehabitatMA’
The following object is masked from ‘package:raster’:
Loading required package: adehabitatHR
Loading required package: deldir
deldir 0.1-15
Loading required package: adehabitatLT
Loading required package: CircStats
Loading required package: MASS
Attaching package: ‘MASS’
The following objects are masked from ‘package:raster’:
Loading required package: boot
> enfa_result install.packages(«sp»)
Error in install.packages : Updating loaded packages
Restarting R session.
Loading required package: raster
Loading required package: sp
Error: package or namespace load failed for ‘raster’ in .doLoadActions(where, attach):
error in load action .__A__.1 for package raster: loadModule(module = «spmod», what = TRUE, env = ns, loadNow = TRUE): Unable to load module «spmod»: object of type ‘closure’ is not subsettable
Error in .requirePackage(package) :
unable to find required package ‘raster’
> install.packages(«sp»)
Error in install.packages : Updating loaded packages
Restarting R session.
Loading required package: raster
Loading required package: sp
Error: package or namespace load failed for ‘raster’ in .doLoadActions(where, attach):
error in load action .__A__.1 for package raster: loadModule(module = «spmod», what = TRUE, env = ns, loadNow = TRUE): Unable to load module «spmod»: object of type ‘closure’ is not subsettable
Error in .requirePackage(package) :
unable to find required package ‘raster’
> install.packages(«sp»)
Error in install.packages : Updating loaded packages
> install.packages(«sp»)
Warning in install.packages :
package ‘sp’ is in use and will not be installed
Perhaps I should just reinstall R? I’ve also noticed it doesn’t detect that I am running Pro 2.2.4; instead it seems to think I am running 1.0.1.232 according to the console.
Источник
Credibly Curious
Nick Tierney’s (mostly) rstats blog
2020-05-29
Error in loadNamespace(Name) : There is No Package Called ‘here’
Nicholas Tierney
This error (or a variant of it) is quite common when using R:
Another variant is:
Let’s list out some ways that you can address this issue.
Install the package
Install the package that is claimed not to be there. That is, for this error:
You install the PKG package (use your package name intead of PKG ):
Why does that work?
is given because R is looking for a package to use, and it cannot find that package. Installing your package means that R can find it, and load it so you can use it!
Another Error: Error in install.packages : object ‘here’ not found
This happens when you write:
You need to write the package that you want to install in quotes:
Why? Well R thinks here is an object, but it requires the R package to be in quotes.
Now, that might not feel like the best reason — the truth is it is to do with a thing called Non Standard Evaluation (NSE), but going into more detail than that is beyond the scope of this blog post.
What I would suggest is this, internalise:
When installing R packages, put the package in quotes: “package”
Another error: Warning in install.packages : package ‘emo’ is not available (for R version 3.6.2)
This can happen if you write:
Why? Well, it could be one of the following below errors:
- Package name misspelt
- Package might not exist on CRAN
It is quite likely that it is not to do with your version of R.
Problem solving: Is the package perhaps misspelt?
I have, more often than I care to admit, had a spelling mistake that caused me to go on a rabbit hole.
Problem solving: Does the package exist on CRAN?
Type “PKG CRAN rstats” into a search engine
Perhaps you might find the right spelling, in which case, install the package with the right spelling using install.packages .
Perhaps you might find that it is on github (or bitbucket or gitlab), not on CRAN.
Let’s take a github example. You need to install an R package from github with a different command.
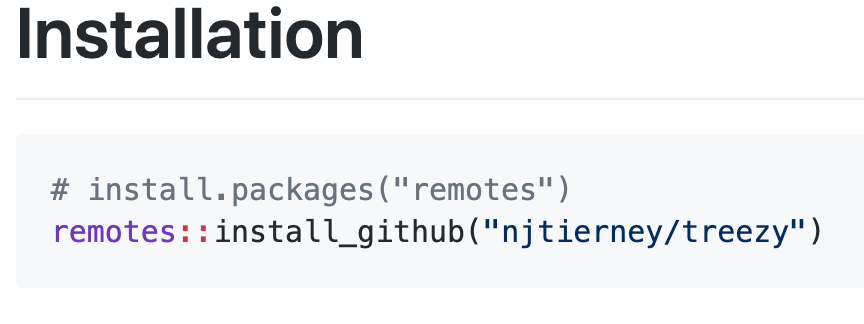
Let’s say we want to install the “treezy” package from github. You scroll down and find the instructions here:
So you will need to do two things:
- Install remotes from CRAN ( install.packages(«remotes» )
- Run remotes::install_github()
Similarly there are packages for R packages that you might find on other repositories such as gitlab ( install_gitlab ) or bitbucket ( install_bitbucket ).
Why write this blog post?
I teach an introduction to data analysis class, and many students encounter this error:
but they do not have the skills and experience to identify how to solve this problem. In class, I decided to showcase how I would try to solve this problem, live, on zoom, to my class. So I googled the error, and then I discovered that the top hit isthis stack overflow page, which was decidedly not helpful for the problem that my students had.
So I wrote this blogpost.
Hopefully it’s helpful!
Next up in this series is tackling this problem:
Thanks
Thanks to Emi Tanaka and Miles McBain for their suggestions on a few helpful additions to the blog post!
Источник
I use R3.3.2. when I want to install the package «arules» , I get this error:
install.packages("arules")
Error in install.packages : Updating loaded packages
Restarting R session...
Loading required package: arules
Loading required package: Matrix
Attaching package: ‘arules’
The following objects are masked from ‘package:base’:
abbreviate, write
Loading required package: twitteR
Loading required package: wavelets
> install.packages("arules")
Error in install.packages : Updating loaded packages
Warning messages:
1: package ‘arules’ was built under R version 3.3.3
2 : package ‘twitteR’ was built under R version 3.3.3
> install.packages("arules")
Installing package into ‘C:/Users/Atapour/Documents/R/win-library/3.3’
(as ‘lib’ is unspecified)
Warning in install.packages :
package ‘arules’ is in use and will not be installed
whenever I atempt to install the «arules» , «matrix» ,»twitteR» , «FactoMineR» and so on, I get the error of :e.g. package ‘twitteR’ was built under R version 3.3.3
I try downloading from cran site(https://cran.r-project.org/web/packages/FactoMineR/index.html) but doest’nt help me anymore.
what should I do to fix it?
I’ve been experiencing this issue as well (and I am an ardent believer of EIKIFJB, so it’s not because I have a stale R environment being loaded), but I think I know the issue on my end at the very least: It’s because of custom Rmarkdown site generators.
Context
I am currently beta testing a site generator (https://github.com/carpentries/sandpaper) with folks and am finding that this loop happens whenever someone tries to update this package inside the project in RStudio. I found this to be true of RStudio cloud projects as well. This has been observed on Ubuntu Linux 20.04 and MacOS
Symptoms
If I inspect the sessionInfo() for different projects after loading them and reloading them with a clean slate, I notice that the context-specific engines are loaded:
Example: blogdown
Packages needed for blogdown are loaded because of the site directive in the index.Rmd file
usethis::create_from_github("zkamvar/zkamvar")
> sessionInfo()
R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_4.1.2 bookdown_0.24 fastmap_1.1.0 cli_3.2.0 htmltools_0.5.2
[6] tools_4.1.2 yaml_2.3.4 rmarkdown_2.11 blogdown_1.7 knitr_1.37
[11] xfun_0.29 digest_0.6.29 rlang_1.0.1 evaluate_0.14
Example: sandpaper
Packages needed for sandpaper are loaded because of the site directive in the index.md file
usethis::create_from_github("carpentries/sandpaper-docs")
> sessionInfo()
R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] ps_1.6.0 rprojroot_2.0.2 digest_0.6.29 assertthat_0.2.1
[5] R6_2.5.1 evaluate_0.14 rlang_1.0.1 cli_3.2.0
[9] fs_1.5.2 sandpaper_0.1.6 callr_3.7.0 rmarkdown_2.11
[13] tools_4.1.2 xfun_0.29 yaml_2.3.4 fastmap_1.1.0
[17] compiler_4.1.2 processx_3.5.2 htmltools_0.5.2 knitr_1.37
One of the primary reasons for R’s popularity is its extensive package ecosystem. On R’s main package repository CRAN alone you have over 10,000 packages available to choose from. Yet, when you first install R you only get a very limited set of core packages “out of the box”. Any further packages that you’d like to use you have to install yourself. How to do that is the topic of this article.
Installing packages from the Comprehensive R Archive Network (CRAN) couldn’t be easier! Simply type install.packages() with the name of your desired package in quotes as first argument. In case you want to install the {dplyr} package you would need to type the following.
install.packages("dplyr")This will install {dplyr} along with all its dependencies, i.e. other packages that {dplyr} uses internally.
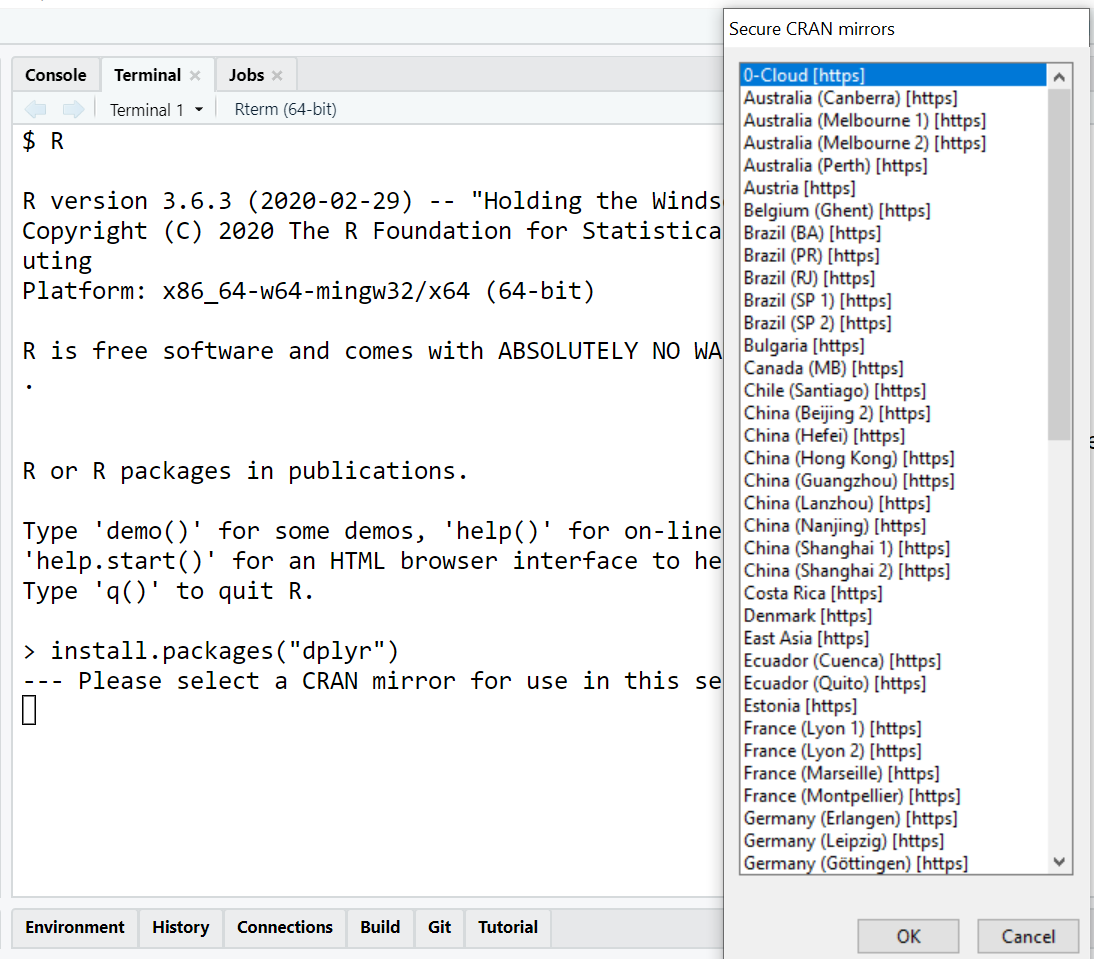
When you execute install.packages() outside of RStudio, e.g. in R’s built-in GUI or in the terminal, you will be asked to select a CRAN mirror.
CRAN is hosted on over 50 different servers spread across the world. If you are lazy you can just select the first entry (0-Cloud) which is the mirror hosted by RStudio. Otherwise choose the mirror nearest to your current location to maximize the download speed.
Installing from Bioconductor
In order to install R packages from Bioconductor—a repository specifically designed for bioinformatics packages—you first need to install the {BiocManager} package from CRAN.
install.packages("BiocManager")Once {BiocManager} has been successfully installed, you can install any package from Bioconductor using the BiocManager::install() function, e.g.
BiocManager::install("ArrayTools")Installing from GitHub
While CRAN is still by far the most popular repository for R packages, you will find quite a lot of packages that are only available from GitHub. Furthermore, if you’d liked to try out the latest development versions of popular packages such as {ggplot2} and {tidyr} you will have to install them from GitHub.
Before you can install any package from GitHub, you need to install the {remotes} package from CRAN.
install.packages("remotes")Now you can install any package from GitHub by providing "username/repository" as argument to remotes::install_github(). For example, to install the latest development version of {ggplot2} from GitHub run this command.
remotes::install_github("tidyverse/ggplot2")Please note that installing a package from GitHub means that you have to install it from source. This requires a proper development environment including (at the minimum) a C and FORTRAN compiler. On Linux and macOS that’s a given but on Windows it’s not. More on that below where I discuss the difference between installing binaries vs. installing from source.
Installing from Other Sources
The {remotes} package can install R packages from many other sources including Gitlab, SVN and Bitbucket as well as your local PC. Here’s a list of all install_*() functions the package contains.
grep(
pattern = "^install_",
x = getNamespaceExports("remotes"),
value = TRUE
)## [1] "install_gitlab" "install_url" "install_github"
## [4] "install_dev" "install_git" "install_version"
## [7] "install_bioc" "install_deps" "install_bitbucket"
## [10] "install_cran" "install_local" "install_svn"
Installing Specifc Package Versions
By default, install.packages() and co. install the latest version of a package. In general that’s a good thing. The most recent version likely incorporates bug fixes for issues from earlier version plus new features. However, there are situations in which you don’t want to install the latest version of a package because it could break your existing code.
The first way to install a specific version of a package is using remotes::install_version(). In addition to the name of the package you’d like to install you pass it a version string as second argument. The function will go though the CRAN archives and install that specific version—if it exists.
remotes::install_version("dplyr", "0.8.5")Another approach to installing a specific package version—or rather the version from a specific point in time—is to make use to use of MRAN’s (Microsoft R Archive Network) daily CRAN snapshots. Every day new packages and new versions of existing packages are uploaded to CRAN. Thus, running install.packages("dplyr") next week might install a different version than running the command today. By using a MRAN snapshot you make sure that whenever you execute the installation command you always get the packages from the date of the snapshot.
MRAN snapshots are URLs with the canonical form https://cran.microsoft.com/snapshot/yyyy-mm-dd/, e.g. https://cran.microsoft.com/snapshot/2020-01-20/. To use such a snapshot you have two options. The first one is to explicitly provide the URL as argument to the repos parameter of install.packages().
install.packages("dplyr", repos = "https://cran.microsoft.com/snapshot/2017-03-15/")Alternatively, you can set the repos option globally.
options(repos = "https://cran.microsoft.com/snapshot/2017-03-15/")Any subsequent call to install.packages() will make use of this globally set option unless you overwrite it by passing another value to the repos parameter.
Binary vs. Source Installation
The install.packages() function has a type parameter that controls whether a package is installed from pre-complied binaries or from source. The former is the default on Windows and some macOS versions while on Linux it’s the latter. So what’s the difference?
When type = "binary", install.packages() will attempt to download a pre-compiled version of the requested package for the operating system (OS) you are working on. Pre-complied means that the package has been complied on another machine with the same OS as yours and subsequently uploaded to CRAN. This is very convenient because not having to compile packages yourself has a huge advantage: it’s (much) faster. If you’ve ever tried to install the {tidyverse} from source you will know what I mean. It’s no overstatement when I say that you should take a long coffee break after entering install.packages("tidyverse", type = "source").
Are there advantages to installing packages from source? In theory yes. You could set certain compiler flags to optimize the code and link to more performant libraries, e.g. multi-threaded BLAS/LAPACK linear algebra libraries. However, that is provided you know what you are doing. I assume that 99.9% of my readers don’t and I include myself to that list. So in short, install from pre-compiled binaries unless they are not available for your OS.
Where Are Packages Installed To?
R has what I like to call the package search path. This is a list of directories in which it looks for R packages when you use library(). You can view these directories using the .libPaths() function.
## [1] "C:/Users/neitmant/AppData/Roaming/R-3.6.3/library"
As you can see on my machine the package search path is just a single directory. That is typically the case on Windows if you install R only for a specific user. If you install it for all users on the machine you usually have two entries: a global one containing all the default packages that ship with R and a user specific one where all additional packages are installed to.
You can add any directory you like to the package search path yourself. Importantly, R will install a new package to the very first entry of the package search path, i.e. .libPaths()[1L].
if (!dir.exists("./library")) {
dir.create("./library")
}
old_libraries <- .libPaths()
.libPaths(c("./library", old_libraries))
.libPaths()## [1] "C:/Users/neitmant/Documents/blog2/library"
## [2] "C:/Users/neitmant/AppData/Roaming/R-3.6.3/library"
From now on any time I call install.packages() the package will be installed in the C:/Users/neitmant/Documents/blog2/library directory rather then the standard C:/Users/neitmant/AppData/Roaming/R-3.6.3/library directory.
Troubleshooting Common Installation Errors
Package ‘abc’ Is Not Available (for R Version x.y.z)
A rather common issue that pops up when trying to install a package from CRAN is package ‘abc’ is not available (for R version x.y.z). On first sight this looks as though the package you requested is not available for the R version you are using (but potentially other ones). In reality the most common reason for this warning is that you misspelled a package name, e.g. you tried to install {gplot2} rather than {ggplot2}.
install.packages("gplot2")## Installing package into 'C:/Users/neitmant/Documents/blog2/library'
## (as 'lib' is unspecified)
## Warning: package 'gplot2' is not available (for R version 3.6.3)
Another reason for this warning could be that the package is not on CRAN (yet). This is often the case with experimental new packages that you may have read about on #RStats twitter. The package will most likely be available on GitHub and can be installed using remotes::install_github().
Yet another potential source for this issue is that you attempted to install a Bioconductor package with install.packages(). For that you have to use BiocManager::install() rather then install.packages().
Updating Loaded Packages
This is a very annoying error. The error message itself is rather self-explanatory. You have already called library() with one of the packages you are attempting to install. Often, though, it’s not obvious which package is causing the problem. You may have called library(dplyr) at the start of your session and are now trying to install another package that depends on {dplyr}. This package likely requires a specific version of {dplyr}. You might have version 0.8.1 installed but 1.0.0 is required. In such cases R will automatically try to install the most recent version of {dplyr}. But if that package is already loaded you’ll get the Updating loaded packages error.
When you encounter this error the first thing you should do is restart your R session and try to install again without loading any packages. If you still get the same error you likely have a library(pkg) call in your .Rprofile file or you load saved objects into your workspace when starting R. I’d highly recommend to do neither of those. As a next step I’d recommend to close any currently active R session. Then go to your terminal—or CMD on Windows—and type R --vanilla. This will start an R session that will ignore your .Rprofile and doesn’t restore any saved objects. You are starting from a clean state so to say. Installing the package should now work.
There Are Binary Versions Available but the Source Versions Are Later
This is a common error when using R on Windows. It is typically followed by the seemingly innocent question: Do you want to install from sources the package which needs compilation?. Unless you have a proper development environment on your Windows machine, i.e. you installed the Rtools software, you should say no. Attempting to install from source when you don’t have a proper development environment is guaranteed to lead to failing package installations.
When you do answer no R will install the previous version of the package for which pre-complied binaries are available rather than the latest version which is only available from source. This is typically the case in the first few days after a new package version has been published on CRAN. Should that still not work try to explicitly install the previous version of the package using remotes::install_version() as described above.
Failed to Create Lock Directory
This error occurs when your last package installation attempt has interrupted abnormally, e.g. when you hit Ctrl-C to terminate it.
Installing package(s) into ‘C:/Users/neitmant/AppData/Roaming/R-3.6.3/library’ (as ‘lib’ is unspecified)
trying URL 'http://cran.us.r-project.org/src/contrib/Rcpp_0.10.2.tar.gz'
Content type 'application/x-gzip' length 2380089 bytes (2.3 Mb)
...
Warning in dir.create(lockdir, recursive = TRUE) :
cannot create dir '/home', reason 'Permission denied'
ERROR: failed to create lock directory ‘C:/Users/neitmant/AppData/Roaming/R-3.6.3/library/00LOCK-Rcpp’
...
According to the install.packages() documentation “[the lock directory] has two purposes: it prevents any other process installing into that library concurrently, and is used to store any previous version of the package to restore on error”.
To mitigate this error you can either manually delete the left-over 00LOCK directory or you tell R to not create one during installation. To delete the 00LOCK directory you can either use the file explorer of your OS or you do it directly from within R using the following command.
unlink("C:/Users/neitmant/AppData/Roaming/R-3.6.3/library/00LOCK-Rcpp", recursive = TRUE)Make sure to pass the exact path you received in the error message as first argument to unlink().
To make install.packages() not create a 00LOCK directory, pass the "--no-lock" flag to its INSTALL_opts parameter.
install.packages("Rcpp", INSTALL_opts = "--no-lock")Wrap Up
Packages are a primary reason for R’s popularity. They can be installed from various sources including—but not limited to—CRAN, Bioconductor and GitHub. While installing packages is most of the time straightforward, errors do occur. This article discussed some of the most common installation errors and provided solutions on how to mitigate them. Now it’s up to you to install your favorite packages and get programming!
Измените ссылку на объект requiredpackages на packages в функции, чтобы устранить логическую ошибку. Поскольку packages — это имя аргумента, содержащего список пакетов, передаваемых функции, это правильный объект для ссылки в цикле for() функции.
Вы также захотите добавить функцию library() в ветку, которая устанавливает пакет, чтобы в конце функции все необходимые пакеты были установлены и загружены.
requiredpackages <- c('ggplot2', 'ggthemes')
install_load <- function(packages){
for (p in packages) {
if (p %in% rownames(installed.packages())) {
library(p, character.only=TRUE)
} else {
install.packages(p)
library(p,character.only = TRUE)
}
}
}
install_load(requiredpackages)
… и вывод:
> install_load(requiredpackages)
Want to understand how all the pieces fit together? See the R for Data Science book:
http://r4ds.had.co.nz/
trying URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.5/ggthemes_4.0.1.tgz'
Content type 'application/x-gzip' length 425053 bytes (415 KB)
==================================================
downloaded 415 KB
The downloaded binary packages are in
/var/folders/2b/bhqlk7xs4712_5b0shwgtg840000gn/T//Rtmp1yfVvz/downloaded_packages
>
Мы также можем использовать sessionInfo(), чтобы подтвердить, что пакеты ggplot2 и ggthemes были загружены в память:
> sessionInfo()
R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14.2
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggthemes_4.0.1 ggplot2_3.1.0
...sessionInfo() output continues.
I figured out that I must have been making a silly mistake in identifying the variable ‘data_path’. I see what I was doing wrong there now.
However, I have run into a different problem: Step 1 under the section Produce a habitat suitability map. When I attempt to load the sp package, R asks me to restart because the package is already currently loaded (it apparently loaded when I used library(raster) earlier in the session). If I click Yes to restart R I end up in a loop where R keeps wanting to restart (and interestingly it also gives a Windows message that it has stopped working, though R stays open i.e. does not crash). If I click No or Cancel, I simply cannot load the sp package.
Below is a copy/paste of my RStudio console (at the end I attempted to let R restart twice by clicking Yes, then I clicked No).
> library(arcgisbinding)
*** Please call arc.check_product() to define a desktop license.
> arc.check_product()
product: ArcGIS Pro ( 12.2.0.12813 )
license: Advanced
version: 1.0.1.232
> data_path <- arc.open(«G:/ArcGIS_LearnLessons/African-Buffalo/LL Ecological Niche Factor Analysis.gdb/ENFA_Environmental_Buffalo_Attributes»)
> install.packages(‘raster’)
trying URL ‘https://cran.rstudio.com/bin/windows/contrib/3.5/raster_2.8-4.zip‘
Content type ‘application/zip’ length 3769843 bytes (3.6 MB)
downloaded 3.6 MB
package ‘raster’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:UserskflammAppDataLocalTempRtmpM9T3Padownloaded_packages
> library(raster)
Loading required package: sp
> arc_raster <- arc.raster(data_path)
> r_raster <- as.raster(arc_raster)
> variables <- as.data.frame(arc_raster$pixel_block())
> variables$pixel_index <- 1:nrow(variables)
> variables <- na.omit(variables)
> pixel_index <- variables$pixel_index
> variables$pixel_index<-NULL
> ecological_variables <-variables[,-56]
> buffalo_presence <- variables[, 56]
> install.packages(«ade4»)
trying URL ‘https://cran.rstudio.com/bin/windows/contrib/3.5/ade4_1.7-13.zip‘
Content type ‘application/zip’ length 5201783 bytes (5.0 MB)
downloaded 5.0 MB
package ‘ade4’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:UserskflammAppDataLocalTempRtmpM9T3Padownloaded_packages
> library(ade4)
> dudi_obj <- dudi.pca(ecological_variables, scannf = FALSE, nf = 10)
> install.packages(«adehabitatHS»)
trying URL ‘https://cran.rstudio.com/bin/windows/contrib/3.5/adehabitatHS_0.3.13.zip‘
Content type ‘application/zip’ length 1515084 bytes (1.4 MB)
downloaded 1.4 MB
package ‘adehabitatHS’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:UserskflammAppDataLocalTempRtmpM9T3Padownloaded_packages
> library(adehabitatHS)
Loading required package: adehabitatMA
Attaching package: ‘adehabitatMA’
The following object is masked from ‘package:raster’:
buffer
Loading required package: adehabitatHR
Loading required package: deldir
deldir 0.1-15
Loading required package: adehabitatLT
Loading required package: CircStats
Loading required package: MASS
Attaching package: ‘MASS’
The following objects are masked from ‘package:raster’:
area, select
Loading required package: boot
> enfa_result <- enfa(dudi_obj, buffalo_presence, scannf = FALSE, nf = 2)
> install.packages(«sp»)
Error in install.packages : Updating loaded packages
Restarting R session…
Loading required package: raster
Loading required package: sp
Error: package or namespace load failed for ‘raster’ in .doLoadActions(where, attach):
error in load action .__A__.1 for package raster: loadModule(module = «spmod», what = TRUE, env = ns, loadNow = TRUE): Unable to load module «spmod»: object of type ‘closure’ is not subsettable
Error in .requirePackage(package) :
unable to find required package ‘raster’
> install.packages(«sp»)
Error in install.packages : Updating loaded packages
Restarting R session…
Loading required package: raster
Loading required package: sp
Error: package or namespace load failed for ‘raster’ in .doLoadActions(where, attach):
error in load action .__A__.1 for package raster: loadModule(module = «spmod», what = TRUE, env = ns, loadNow = TRUE): Unable to load module «spmod»: object of type ‘closure’ is not subsettable
Error in .requirePackage(package) :
unable to find required package ‘raster’
> install.packages(«sp»)
Error in install.packages : Updating loaded packages
> install.packages(«sp»)
Warning in install.packages :
package ‘sp’ is in use and will not be installed
Perhaps I should just reinstall R? I’ve also noticed it doesn’t detect that I am running Pro 2.2.4; instead it seems to think I am running 1.0.1.232 according to the console.
В большинстве случаев, особенно при установке пакетов из CRAN никаких проблем не возникает, но периодически всё таки вы можете столкнуться с некоторыми ошибками.
В этой статье я со временем буду добавлять материал с описанием различных ошибок которые возникают при установке пакетов.
package ‘foo’ is not available”
Достаточно распространённая ошибка, текст которой к сожалению не сообщает о реальной проблеме, которой была эта ошибка вызвана.
В одной из статей на RBloggers, автор опубликовал подробный чек лист как с этой ошибкой бороться.
- Неправильно написанное название пакета. Первое, что стоит проверить — это правильно ли вы написали название пакета, который хотите установить. R регистро чувствительный язык, поэтому даже одна буква написанная не в том регистре будет считаться неправильным именем пакета. Лучший способ копировать название пакета из CRAN.
- Проверьте своё интернет соединение. Звучит банально, но иногда в процессе разработки когда мы можем не заметить, что уже более часа работаем без доступа к интернету.
- Проверьте сеть CRAN. Возможно в момент установки пакета не доступны сервера CRAN, для проверки просто попробуйте загрузить главный сайт проекта. Если основное зеркало CRAN не доступно, вы можете установить пакет из другого, используя аргумент
reposвнутри функцииinstall.packages(). - Убедитесь в том, что пакет был опубликован на CRAN. Перед тем как пакет попадёт на CRAN, зачастую автор проводит большую работу над его тестированием, и доработкой функционала. Как правило на CRAN публикуются пакеты чей код и функционал уже устоялся, и считался стабильным. Поэтому 4ый шаг, просто убедитесь в том, что нужный пакет уже опубликван. Если пакета нет на CRAN, то скорее всего вы можете найти его на GitHub, и установить с помощью команды
devtools::install_github('pkg_name'). - Убедитесь в том, что пакет не был удалён из CRAN. Требования политики публикации пакетов в CRAN не редко меняются, если автор перестал поддерживать свой пакет, и с течением времени этот пакет стал нарушать правила политики публикации CRAN, то его удалят из репозитория. В таком случае перейдя на страницу пакета https://cran.r-project.org/package=PACKAGE_NAME, вы увидите сообщение “Package ‘foo’ was removed from the CRAN repository.”, что и свидетельствует об удалении пакета из CRAN. Если вы получите ошибку 404, значит пакет ранее не был опубликован на CRAN.
unable to create temporary directory
Недавно столкнулся с проблемой при установке пакетов из RStudio — ОШИБКА: нет прав для установочной папки ‘C:/Users/Documents/R/win-library/3.5’.
Та же ошибка может возвращать и следующие сообщения:
- ‘lib = «C:/Users/Alsey/Documents/R/win-library/3.5″‘ is not writable
- Error in install.packages : unable to create temporary directory ‘C:UsersAlseyDocumentsRwin-library3.5file34ac62294fbd’
С чем связано её появление я так и не понял, но устранить получилось следующим образом:
- Создайте новую папку, которая будет в последствии вашей пользовательской библиотекой R пакетов, лучше создавать её не на диске C, что бы в последствии не сталкиваться с такой же проблемой.
- В основной вашей рабочей директории (скорее всего это папка ‘Документы’ которая обычно находится в каталоге пользователя Windows, примерный путь к ней C:UsersAlseyDocuments ), найдите файл .Renviron, на данный момент он будет пустой, это файл конфигурации R, именно в нём можно менять дефолтные значения глобальных переменных R, вам необходимо в него вписать одну строку R_LIBS_USER=D:/r_library, где D:/r_library это путь к созданной на первом шаге папке.
- Далее перезапустите сессию R, и можете убедиться с помощью команды
.libPaths(), что созданная на первом шаге папка стала пользовательской библиотекой для установки и хранения пакетов.
Could not find tools necessary to compile a package
С этой ошибкой я столкнулся при установке пакетов из GitHub после обновления R до более новой версии.
Ответ я нашел вот тут.
Проблема возникает при попытке RStudio проверить установленные у вас инструменты сборки пакетов, для того, что бы подвить эту проверку необходимо перед установкой пакета установить следующую опцию.
options(buildtools.check = function(action) TRUE )После чего можно устанавливать пакет.
Found continuation line starting ‘ shortcut functio …’ at begin of record
Эта проблема появилась при установке пакетов из GitHub с помоью devtools не так давно, и связана она с файлом DESCRIPTION.
Возникает в ситуации когда в файле DESCRIPTION имеется многострочное описание в поле » Description: «.
Для исправления вам необходимо форкнуть нужный пакет на GitHub.
И сделать однострочным поле » Description: » в файле DESCRIPTION, далее устанавливайте пакет уже из своего репозитория.
(converted from warning) installation of package ‘C:/Users/Alsey/AppData/Local/Temp/2/Rtmp4g880D/file259c11b85f00/vctrs_0.1.0.9003.tar.gz’ had non-zero exit status
Вызвана данная ошибка конфликтом возникающим при установке пакетов одновременно для разных версий ядра R, 32 и 64 битных.
Полный текст ошибки из консоли:
error: ld returned 1 exit status no DLL was created ERROR: compilation failed for package 'vctrs' * removing 'D:/r_library/vctrs' * restoring previous 'D:/r_library/vctrs' Error in i.p(...) : (converted from warning) installation of package ‘C:/Users/Alsey/AppData/Local/Temp/Rtmpyu30Ew/file35cc2a261c8f/vctrs_0.2.0.9000.tar.gz’ had non-zero exit status
Узнать разрядность версии R в которой вы работаете можно двумя способами:
Sys.getenv("R_ARCH")
[1] "/x64"
В случае если вы используете 32 битный R вернётся значение "/i386".
Sys.info()[["machine"]] [1] "x86-64"
На 32 битном R вы получите "x86_32".
После того, как мы определили разрядность ядра требуется пойти одним из описанных способов, для 64 битной версии просто используйте при установке пакета опцию "--no-multiarch".
devtools::install_github("username/repos", INSTALL_opts = "--no-multiarch")
Если у вас 32 битная версия, то необходимо изменить в переменной окружения PATH путь к утилите RTools с C:Rtoolsmingw_64bin на C:Rtoolsmingw_32bin. О том как это сделать можно узнать в этой статье .
Далее запускаем установку пакета только для 32 битной версии с помощью опции "--no-multiarch", так же как и ранее было показано с примером для 64 разрядного R.
Warning Message: cannot remove prior installation of package ‘X’
Если вы увидели такое сообщение при попытке установить какой-либо пакет, это говорит о том, что пакет в данный момент заблокирован каким – то процессом. Первое, что вам необходимо попробовать сделать это перезапустить RStudio, и попробовать повторно запустить установку пакета.
Если перезапуск RStudio не помог, то откройте диспетчер задач, и на вкладке подробности посмотрите, нет ли у вас каких либо зависших R сеансов.
Если такие есть их необходимо завершить, и попробовать повторно запустить процесс установки пакета.
Если и это не помогло, то идём третьим способом.
- Найдите путь к папкам, в которых у вас установлены пакеты, делается это командой
.libPaths(). - В ручном режиме удалите папку с пакетом, который пытаетесь установить.
- Откройте RStudio и повторите попытку установить пакет.
Один из перечисленных выше способов должен сработать.
Статья будет постоянно дополняться, дата последнего редактирования 6 октября 2020 года.
Я использую R3.3.2. когда я хочу установить пакет «arules», я получаю эту ошибку:
install.packages("arules")Error in install.packages : Updating loaded packages
Restarting R session...
Loading required package: arules
Loading required package: MatrixAttaching package: ‘arules
The following objects are masked from ‘package:base:
abbreviate, write
Loading required package: twitteR
Loading required package: wavelets
> install.packages("arules")
Error in install.packages : Updating loaded packages
Warning messages:
1: package ‘arules was built under R version 3.3.3
2 : package ‘twitteR was built under R version 3.3.3
> install.packages("arules")
Installing package into ‘C:/Users/Atapour/Documents/R/win-library/3.3
(as ‘lib is unspecified)
Warning in install.packages :
package ‘arules is in use and will not be installed
всякий раз, когда я пытаюсь установить «arules», «matrix», «twitteR», «FactoMineR» и т.д., я получаю ошибку: например, пакет «twitteR» был создан под R версии 3.3.3
Я пытаюсь загрузить с сайта cran (https://cran.r-project.org/web/packages/FactoMineR/index.html), но doest’nt мне больше не помогает. что я должен сделать, чтобы исправить это?