In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
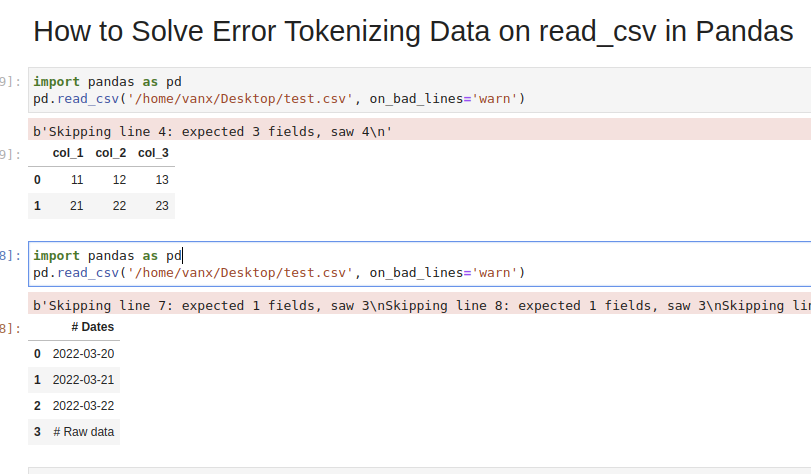
col_1,col_2,col_3
11,12,13
21,22,23
31,32,33,44
which we are going to read by — read_csv() method:
import pandas as pd
pd.read_csv('test.csv')
We will get an error:
ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines='skip' in order to skip bad lines:
pd.read_csv('test.csv', on_bad_lines='skip')
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
pd.read_csv('test.csv', on_bad_lines='warn')
warning like:
b'Skipping line 4: expected 3 fields, saw 4n'
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
sep—lineterminator—engine
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
import pandas as pd
pd.read_csv('test.csv', sep='t', lineterminator='rn')
Note that delimiter is an alias for sep.
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
cpythonpyarrow
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
pd.read_csv('test.csv', engine='python')
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
pd.read_csv('test.csv', skiprows=1, engine='python')
or skipping the last lines:
pd.read_csv('test.csv', skipfooter=1, engine='python')
Note that we need to use python as the read_csv() engine for parameter — skipfooter.
In some cases header=None can help in order to solve the error:
pd.read_csv('test.csv', header=None)
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
Dates
2022-03-20
2022-03-21
2022-03-22
Raw data
col_1,col_2,col_3
11,12,13
21,22,23
31,32,33
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
df_temp = pd.read_csv('test.csv', sep='@@', engine='python', names=['col'])
ix_last_start = df_temp[df_temp['col'].str.contains('# Raw data')].index.max()
result is:
4
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
df_analytics = pd.read_csv(file, skiprows=ix_last_start)
Conclusion
In this post we saw how to investigate the error:
ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
I’m trying to use pandas to manipulate a .csv file but I get this error:
pandas.parser.CParserError: Error tokenizing data. C error: Expected 2 fields in line 3, saw 12
I have tried to read the pandas docs, but found nothing.
My code is simple:
path = 'GOOG Key Ratios.csv'
#print(open(path).read())
data = pd.read_csv(path)
How can I resolve this? Should I use the csv module or another language ?
File is from Morningstar (http://financials.morningstar.com/ratios/r.html?t=GOOG®ion=usa&culture=en-US)
you could also try;
data = pd.read_csv('file1.csv', on_bad_lines='skip')
Do note that this will cause the offending lines to be skipped.
It might be an issue with
- the delimiters in your data
- the first row, as @TomAugspurger noted
To solve it, try specifying the sep and/or header arguments when calling read_csv. For instance,
df = pandas.read_csv(filepath, sep='delimiter', header=None)
In the code above, sep defines your delimiter and header=None tells pandas that your source data has no row for headers / column titles. Thus saith the docs: «If file contains no header row, then you should explicitly pass header=None». In this instance, pandas automatically creates whole-number indices for each field {0,1,2,…}.
According to the docs, the delimiter thing should not be an issue. The docs say that «if sep is None [not specified], will try to automatically determine this.» I however have not had good luck with this, including instances with obvious delimiters.
Another solution may be to try auto detect the delimiter
# use the first 2 lines of the file to detect separator
temp_lines = csv_file.readline() + 'n' + csv_file.readline()
dialect = csv.Sniffer().sniff(temp_lines, delimiters=';,')
# remember to go back to the start of the file for the next time it's read
csv_file.seek(0)
df = pd.read_csv(csv_file, sep=dialect.delimiter)
The parser is getting confused by the header of the file. It reads the first row and infers the number of columns from that row. But the first two rows aren’t representative of the actual data in the file.
Try it with data = pd.read_csv(path, skiprows=2)
This is definitely an issue of delimiter, as most of the csv CSV are got create using sep='/t' so try to read_csv using the tab character (t) using separator /t. so, try to open using following code line.
data=pd.read_csv("File_path", sep='t')
Your CSV file might have variable number of columns and read_csv inferred the number of columns from the first few rows. Two ways to solve it in this case:
1) Change the CSV file to have a dummy first line with max number of columns (and specify header=[0])
2) Or use names = list(range(0,N)) where N is the max number of columns.
I had this problem, where I was trying to read in a CSV without passing in column names.
df = pd.read_csv(filename, header=None)
I specified the column names in a list beforehand and then pass them into names, and it solved it immediately. If you don’t have set column names, you could just create as many placeholder names as the maximum number of columns that might be in your data.
col_names = ["col1", "col2", "col3", ...]
df = pd.read_csv(filename, names=col_names)
I had this problem as well but perhaps for a different reason. I had some trailing commas in my CSV that were adding an additional column that pandas was attempting to read. Using the following works but it simply ignores the bad lines:
data = pd.read_csv('file1.csv', error_bad_lines=False)
If you want to keep the lines an ugly kind of hack for handling the errors is to do something like the following:
line = []
expected = []
saw = []
cont = True
while cont == True:
try:
data = pd.read_csv('file1.csv',skiprows=line)
cont = False
except Exception as e:
errortype = e.message.split('.')[0].strip()
if errortype == 'Error tokenizing data':
cerror = e.message.split(':')[1].strip().replace(',','')
nums = [n for n in cerror.split(' ') if str.isdigit(n)]
expected.append(int(nums[0]))
saw.append(int(nums[2]))
line.append(int(nums[1])-1)
else:
cerror = 'Unknown'
print 'Unknown Error - 222'
if line != []:
# Handle the errors however you want
I proceeded to write a script to reinsert the lines into the DataFrame since the bad lines will be given by the variable ‘line’ in the above code. This can all be avoided by simply using the csv reader. Hopefully the pandas developers can make it easier to deal with this situation in the future.
 Solution
SolutionThe following worked for me (I posted this answer, because I specifically had this problem in a Google Colaboratory Notebook):
df = pd.read_csv("/path/foo.csv", delimiter=';', skiprows=0, low_memory=False)
You can try;
data = pd.read_csv('file1.csv', sep='t')
I came across the same issue. Using pd.read_table() on the same source file seemed to work. I could not trace the reason for this but it was a useful workaround for my case. Perhaps someone more knowledgeable can shed more light on why it worked.
Edit:
I found that this error creeps up when you have some text in your file that does not have the same format as the actual data. This is usually header or footer information (greater than one line, so skip_header doesn’t work) which will not be separated by the same number of commas as your actual data (when using read_csv). Using read_table uses a tab as the delimiter which could circumvent the users current error but introduce others.
I usually get around this by reading the extra data into a file then use the read_csv() method.
The exact solution might differ depending on your actual file, but this approach has worked for me in several cases
I’ve had this problem a few times myself. Almost every time, the reason is that the file I was attempting to open was not a properly saved CSV to begin with. And by «properly», I mean each row had the same number of separators or columns.
Typically it happened because I had opened the CSV in Excel then improperly saved it. Even though the file extension was still .csv, the pure CSV format had been altered.
Any file saved with pandas to_csv will be properly formatted and shouldn’t have that issue. But if you open it with another program, it may change the structure.
Hope that helps.
The dataset that I used had a lot of quote marks («) used extraneous of the formatting. I was able to fix the error by including this parameter for read_csv():
quoting=3 # 3 correlates to csv.QUOTE_NONE for pandas
I’ve had a similar problem while trying to read a tab-delimited table with spaces, commas and quotes:
1115794 4218 "k__Bacteria", "p__Firmicutes", "c__Bacilli", "o__Bacillales", "f__Bacillaceae", ""
1144102 3180 "k__Bacteria", "p__Firmicutes", "c__Bacilli", "o__Bacillales", "f__Bacillaceae", "g__Bacillus", ""
368444 2328 "k__Bacteria", "p__Bacteroidetes", "c__Bacteroidia", "o__Bacteroidales", "f__Bacteroidaceae", "g__Bacteroides", ""
import pandas as pd
# Same error for read_table
counts = pd.read_csv(path_counts, sep='t', index_col=2, header=None, engine = 'c')
pandas.io.common.CParserError: Error tokenizing data. C error: out of memory
This says it has something to do with C parsing engine (which is the default one). Maybe changing to a python one will change anything
counts = pd.read_table(path_counts, sep='t', index_col=2, header=None, engine='python')
Segmentation fault (core dumped)
Now that is a different error.
If we go ahead and try to remove spaces from the table, the error from python-engine changes once again:
1115794 4218 "k__Bacteria","p__Firmicutes","c__Bacilli","o__Bacillales","f__Bacillaceae",""
1144102 3180 "k__Bacteria","p__Firmicutes","c__Bacilli","o__Bacillales","f__Bacillaceae","g__Bacillus",""
368444 2328 "k__Bacteria","p__Bacteroidetes","c__Bacteroidia","o__Bacteroidales","f__Bacteroidaceae","g__Bacteroides",""
_csv.Error: ' ' expected after '"'
And it gets clear that pandas was having problems parsing our rows. To parse a table with python engine I needed to remove all spaces and quotes from the table beforehand. Meanwhile C-engine kept crashing even with commas in rows.
To avoid creating a new file with replacements I did this, as my tables are small:
from io import StringIO
with open(path_counts) as f:
input = StringIO(f.read().replace('", ""', '').replace('"', '').replace(', ', ',').replace('',''))
counts = pd.read_table(input, sep='t', index_col=2, header=None, engine='python')
tl;dr
Change parsing engine, try to avoid any non-delimiting quotes/commas/spaces in your data.
Use delimiter in parameter
pd.read_csv(filename, delimiter=",", encoding='utf-8')
It will read.
For those who are having similar issue with Python 3 on linux OS.
pandas.errors.ParserError: Error tokenizing data. C error: Calling
read(nbytes) on source failed. Try engine='python'.
Try:
df.read_csv('file.csv', encoding='utf8', engine='python')
In my case the separator was not the default «,» but Tab.
pd.read_csv(file_name.csv, sep='\t',lineterminator='\r', engine='python', header='infer')
Note: «t» did not work as suggested by some sources. «\t» was required.
I came across multiple solutions for this issue. Lot’s of folks have given the best explanation for the answers also. But for the beginners I think below two methods will be enough :
import pandas as pd
#Method 1
data = pd.read_csv('file1.csv', error_bad_lines=False)
#Note that this will cause the offending lines to be skipped.
#Method 2 using sep
data = pd.read_csv('file1.csv', sep='t')
Although not the case for this question, this error may also appear with compressed data. Explicitly setting the value for kwarg compression resolved my problem.
result = pandas.read_csv(data_source, compression='gzip')
Sometimes the problem is not how to use python, but with the raw data.
I got this error message
Error tokenizing data. C error: Expected 18 fields in line 72, saw 19.
It turned out that in the column description there were sometimes commas. This means that the CSV file needs to be cleaned up or another separator used.
following sequence of commands works (I lose the first line of the data -no header=None present-, but at least it loads):
df = pd.read_csv(filename,
usecols=range(0, 42))
df.columns = ['YR', 'MO', 'DAY', 'HR', 'MIN', 'SEC', 'HUND',
'ERROR', 'RECTYPE', 'LANE', 'SPEED', 'CLASS',
'LENGTH', 'GVW', 'ESAL', 'W1', 'S1', 'W2', 'S2',
'W3', 'S3', 'W4', 'S4', 'W5', 'S5', 'W6', 'S6',
'W7', 'S7', 'W8', 'S8', 'W9', 'S9', 'W10', 'S10',
'W11', 'S11', 'W12', 'S12', 'W13', 'S13', 'W14']
Following does NOT work:
df = pd.read_csv(filename,
names=['YR', 'MO', 'DAY', 'HR', 'MIN', 'SEC', 'HUND',
'ERROR', 'RECTYPE', 'LANE', 'SPEED', 'CLASS',
'LENGTH', 'GVW', 'ESAL', 'W1', 'S1', 'W2', 'S2',
'W3', 'S3', 'W4', 'S4', 'W5', 'S5', 'W6', 'S6',
'W7', 'S7', 'W8', 'S8', 'W9', 'S9', 'W10', 'S10',
'W11', 'S11', 'W12', 'S12', 'W13', 'S13', 'W14'],
usecols=range(0, 42))
CParserError: Error tokenizing data. C error: Expected 53 fields in line 1605634, saw 54
Following does NOT work:
df = pd.read_csv(filename,
header=None)
CParserError: Error tokenizing data. C error: Expected 53 fields in line 1605634, saw 54
Hence, in your problem you have to pass usecols=range(0, 2)
I believe the solutions,
,engine='python'
, error_bad_lines = False
will be good if it is dummy columns and you want to delete it.
In my case, the second row really had more columns and I wanted those columns to be integrated and to have the number of columns = MAX(columns).
Please refer to the solution below that I could not read anywhere:
try:
df_data = pd.read_csv(PATH, header = bl_header, sep = str_sep)
except pd.errors.ParserError as err:
str_find = 'saw '
int_position = int(str(err).find(str_find)) + len(str_find)
str_nbCol = str(err)[int_position:]
l_col = range(int(str_nbCol))
df_data = pd.read_csv(PATH, header = bl_header, sep = str_sep, names = l_col)
As far as I can tell, and after taking a look at your file, the problem is that the csv file you’re trying to load has multiple tables. There are empty lines, or lines that contain table titles. Try to have a look at this Stackoverflow answer. It shows how to achieve that programmatically.
Another dynamic approach to do that would be to use the csv module, read every single row at a time and make sanity checks/regular expressions, to infer if the row is (title/header/values/blank). You have one more advantage with this approach, that you can split/append/collect your data in python objects as desired.
The easiest of all would be to use pandas function pd.read_clipboard() after manually selecting and copying the table to the clipboard, in case you can open the csv in excel or something.
Irrelevant:
Additionally, irrelevant to your problem, but because no one made mention of this: I had this same issue when loading some datasets such as seeds_dataset.txt (http://archive.ics.uci.edu/ml/datasets/seeds) from UCI. In my case, the error was occurring because some separators had more whitespaces than a true tab t. See line 3 in the following for instance
14.38 14.21 0.8951 5.386 3.312 2.462 4.956 1
14.69 14.49 0.8799 5.563 3.259 3.586 5.219 1
14.11 14.1 0.8911 5.42 3.302 2.7 5 1
Therefore, use t+ in the separator pattern instead of t.
data = pd.read_csv(path, sep='t+`, header=None)
An alternative that I have found to be useful in dealing with similar parsing errors uses the CSV module to re-route data into a pandas df. For example:
import csv
import pandas as pd
path = 'C:/FileLocation/'
file = 'filename.csv'
f = open(path+file,'rt')
reader = csv.reader(f)
#once contents are available, I then put them in a list
csv_list = []
for l in reader:
csv_list.append(l)
f.close()
#now pandas has no problem getting into a df
df = pd.DataFrame(csv_list)
I find the CSV module to be a bit more robust to poorly formatted comma separated files and so have had success with this route to address issues like these.
Error tokenizing data. C error: Expected 2 fields in line 3, saw 12
The error gives a clue to solve the problem » Expected 2 fields in line 3, saw 12″, saw 12 means length of the second row is 12 and first row is 2.
When you have data like the one shown below, if you skip rows then most of the data will be skipped
data = """1,2,3
1,2,3,4
1,2,3,4,5
1,2
1,2,3,4"""
If you dont want to skip any rows do the following
#First lets find the maximum column for all the rows
with open("file_name.csv", 'r') as temp_f:
# get No of columns in each line
col_count = [ len(l.split(",")) for l in temp_f.readlines() ]
### Generate column names (names will be 0, 1, 2, ..., maximum columns - 1)
column_names = [i for i in range(max(col_count))]
import pandas as pd
# inside range set the maximum value you can see in "Expected 4 fields in line 2, saw 8"
# here will be 8
data = pd.read_csv("file_name.csv",header = None,names=column_names )
Use range instead of manually setting names as it will be cumbersome when you have many columns.
Additionally you can fill up the NaN values with 0, if you need to use even data length. Eg. for clustering (k-means)
new_data = data.fillna(0)
I had a similar case as this and setting
train = pd.read_csv('input.csv' , encoding='latin1',engine='python')
worked
use
pandas.read_csv('CSVFILENAME',header=None,sep=', ')
when trying to read csv data from the link
http://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data
I copied the data from the site into my csvfile. It had extra spaces so used sep =’, ‘ and it worked 
Simple resolution: Open the csv file in excel & save it with different name file of csv format. Again try importing it spyder, Your problem will be resolved!
I had a dataset with prexisting row numbers, I used index_col:
pd.read_csv('train.csv', index_col=0)
This is what I did.
sep='::' solved my issue:
data=pd.read_csv('C:\Users\HP\Downloads\NPL ASSINGMENT 2 imdb_labelled\imdb_labelled.txt',engine='python',header=None,sep='::')
I have the same problem when read_csv: ParserError: Error tokenizing data.
I just saved the old csv file to a new csv file. The problem is solved!
Comments Section
Stumbled on this answer, is there a way to fill missing columns on lines that outputs something like
expected 8 fields, saw 9?
The better solution is to investigate the offending file and to correct the bad lines so that they can be read by
read_csv. @PetraBarus, why not just add columns to the CSV files that are missing them (with null values as needed)?
Yes, I just did that. It’s much easier by adding columns. Opening CSV in a spreadsheet does this.
@MichaelQueue : This is incorrect. A CSV, although commonly delimited by a comma, may be delimited by other characters as well. See CSV specifications. It may be a comma, a tab (‘t’), semicolon, and possibly additional spaces.

If this error arises when reading a file written by
pandas.to_csv(), it MIGHT be because there is a ‘r’ in a column names, in which case to_csv() will actually write the subsequent column names into the first column of the data frame, causing a difference between the number of columns in the first X rows. This difference is one cause of the C error.
Sometime just explicitly giving the «sep» parameter helps. Seems to be a parser issue.
in my case it was a separator issue. read_csv apparently defaults to commas, and i have text fields which include commas (and the data was stored with a different separator anyway)
This error may arise also when you’re using comma as a delimiter and you have more commas then expected (more fields in the error row then defined in the header). So you need to either remove the additional field or remove the extra comma if it’s there by mistake. You can fix this manually and then you don’t need to skip the error lines.
Passing in
names=["col1", "col2", ...]for the max number of expected columns also works, and this is how I solved this issue when I came across it. See: stackoverflow.com/questions/18039057/…
Comment from gilgamash helped me. Open csv file in a text editor (like the windows editor or notepad++) so see which character is used for separation. If it’s a semicolon e.g. try
pd.read_csv("<path>", sep=";"). Do not use Excel for checking as it sometimes puts the data into columns by default and therefore removes the separator.
stumbled across the exact same thing. As far as I’m concerned, this is the correct answer. The accepted one just hides the error.
I experimented problems when not setting
|as the delimiter for my .csv. I rather to try this approach first, instead of skipping lines, or bad lines.
I also had the same problem, I assumed «t» would be detected as a delimiter by default. It worked when I explicitly set the delimiter to «t».
This should not be the accepted answer, lines will be skipped and you don’t know why…
If commas are used in the values but tab is the delimiter and sep is not used (or as suggested above the delimiters whatever it is assumed to be occurs in the values) then this error will arise. Make sure that the delimiter does not occur in any of the values else some rows will appear to have the incorrect number of columns
I’m using excel 2016 while creating the CSV, and using sep=’;’ work for me
This answer better because the row doesn’t get deleted compared to if using the error_bad_line=False. Additionally, you can easily figure out which lines were the problem ones once making a dataframe from this solution.
I agree with @zipline86. This answer is safe and intelligent.
I will take any better way to find the number of columns in the error message than what i just did
Correct answer for me too. +1
While this code may solve the question, including an explanation (//meta.stackexchange.com/q/114762) of how and why this solves the problem would really help to improve the quality of your post, and probably result in more up-votes. Remember that you are answering the question for readers in the future, not just the person asking now. Please edit your answer to add explanations and give an indication of what limitations and assumptions apply. From Review (/review/late-answers/27117963)
this solution its too hackish to me, but it works. I solved my issue passing engine=’python’ in read_csv to deal with variable columns size
names=range(N)should suffice (usingpandas=1.1.2here)
I had the same problem for a large .csv file (~250MB), with some corrupted lines spanning less columns than the data frame actually has. I was able to avoid the exception in two ways: 1) By modifying (for example deleting) a couple of unrelated rows far away from the line causing the exception. 2) By setting
low_memory=False. In other .csv files with the same type of mal-formatted lines, I don’t observe any problems. In summary, this indicates that the handling of large-file bypandas.read_csv()somehow is flawed.
I filed a bug report related to my previous comment.
had a similar issue. Realized it was due to my csv file having a value with a comma in it. Had to encapsulate it with » »
Thank you for this solution !! It’s a very useful tip.
Dude! Thank you. Your solution worked like a light switch.
Thanks, delimiter=»t+» solved the error for me!
I had a file where there were commas in some certain fields/columns and while trying to read through pandas read_csv() it was failing, but after specifying engine=»python» within read_csv() as a parameter it worked — Thanks for this!
Related Topics
python
pandas
csv
Mentions
Saurabh Tripathi
Dr Manhattan
Ronak Shah
Abuteau
Svenskithesource
Richie
Grisaitis
Tom Augspurger
Lucas
Ajean
Computerist
Steven Rouk
Robert Geiger
Double Beep
Manodhya Opallage
El Pastor
Bhavesh Kumar
Zstack
Regularly Scheduled Programming
Sachin
Kims Sifers
Bcoz
Kareem Jeiroudi
Laurent T
Abhishek Tripathi
Adewole Adesola
Naseer
Amran Hossen
Gogasca
Ssuperczynski
Simin Zuo
Constantstranger
References
18039057/python-pandas-error-tokenizing-data
While reading a CSV file, you may get the “Pandas Error Tokenizing Data“. This mostly occurs due to the incorrect data in the CSV file.
You can solve python pandas error tokenizing data error by ignoring the offending lines using error_bad_lines=False.
In this tutorial, you’ll learn the cause and how to solve the error tokenizing data error.
If you’re in Hurry
You can use the below code snippet to solve the tokenizing error. You can solve the error by ignoring the offending lines and suppressing errors.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', error_bad_lines=False, engine ='python')
dfIf You Want to Understand Details, Read on…
In this tutorial, you’ll learn the causes for the exception “Error Tokenizing Data” and how it can be solved.
Cause of the Problem
- CSV file has two header lines
- Different separator is used
r– is a new line character and it is present in column names which makes subsequent column names to be read as next line- Lines of the CSV files have inconsistent number of columns
In the case of invalid rows which has an inconsistent number of columns, you’ll see an error as Expected 1 field in line 12, saw m. This means it expected only 1 field in the CSV file but it saw 12 values after tokenizing it. Hence, it doesn’t know how the tokenized values need to be handled. You can solve the errors by using one of the options below.
Finding the Problematic Line (Optional)
If you want to identify the line which is creating the problem while reading, you can use the below code snippet.
It uses the CSV reader. hence it is similar to the read_csv() method.
Snippet
import csv
with open("sample.csv", 'rb') as file_obj:
reader = csv.reader(file_obj)
line_no = 1
try:
for row in reader:
line_no += 1
except Exception as e:
print (("Error in the line number %d: %s %s" % (line_no, str(type(e)), e.message)))Using Err_Bad_Lines Parameter
When there is insufficient data in any of the rows, the tokenizing error will occur.
You can skip such invalid rows by using the err_bad_line parameter within the read_csv() method.
This parameter controls what needs to be done when a bad line occurs in the file being read.
If it’s set to,
False– Errors will be suppressed for Invalid linesTrue– Errors will be thrown for invalid lines
Use the below snippet to read the CSV file and ignore the invalid lines. Only a warning will be shown with the line number when there is an invalid lie found.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', error_bad_lines=False)
dfIn this case, the offending lines will be skipped and only the valid lines will be read from CSV and a dataframe will be created.
Using Python Engine
There are two engines supported in reading a CSV file. C engine and Python Engine.
C Engine
- Faster
- Uses C language to parse the CSV file
- Supports
float_precision - Cannot automatically detect the separator
- Doesn’t support skipping footer
Python Engine
- Slower when compared to C engine but its feature complete
- Uses Python language to parse the CSV file
- Doesn’t support
float_precision. Not required with Python - Can automatically detect the separator
- Supports skipping footer
Using the python engine can solve the problems faced while parsing the files.
For example, When you try to parse large CSV files, you may face the Error tokenizing data. c error out of memory. Using the python engine can solve the memory issues while parsing such big CSV files using the read_csv() method.
Use the below snippet to use the Python engine for reading the CSV file.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', engine='python', error_bad_lines=False)
dfThis is how you can use the python engine to parse the CSV file.
Optionally, this could also solve the error Error tokenizing data. c error out of memory when parsing the big CSV files.
Using Proper Separator
CSV files can have different separators such as tab separator or any other special character such as ;. In this case, an error will be thrown when reading the CSV file, if the default C engine is used.
You can parse the file successfully by specifying the separator explicitly using the sep parameter.
As an alternative, you can also use the python engine which will automatically detect the separator and parse the file accordingly.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', sep='t')
dfThis is how you can specify the separator explicitly which can solve the tokenizing errors while reading the CSV files.
Using Line Terminator
CSV file can contain r carriage return for separating the lines instead of the line separator n.
In this case, you’ll face CParserError: Error tokenizing data. C error: Buffer overflow caught - possible malformed input file when the line contains the r instead on n.
You can solve this error by using the line terminator explicitly using the lineterminator parameter.
Snippet
df = pd.read_csv('sample.csv',
lineterminator='n')This is how you can use the line terminator to parse the files with the terminator r.
Using header=None
CSV files can have incomplete headers which can cause tokenizing errors while parsing the file.
You can use header=None to ignore the first line headers while reading the CSV files.
This will parse the CSV file without headers and create a data frame. You can also add headers to column names by adding columns attribute to the read_csv() method.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, error_bad_lines=False)
dfThis is how you can ignore the headers which are incomplete and cause problems while reading the file.
Using Skiprows
CSV files can have headers in more than one row. This can happen when data is grouped into different sections and each group is having a name and has columns in each section.
In this case, you can ignore such rows by using the skiprows parameter. You can pass the number of rows to be skipped and the data will be read after skipping those number of rows.
Use the below snippet to skip the first two rows while reading the CSV file.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, skiprows=2, error_bad_lines=False)
dfThis is how you can skip or ignore the erroneous headers while reading the CSV file.
Reading As Lines and Separating
In a CSV file, you may have a different number of columns in each row. This can occur when some of the columns in the row are considered optional. You may need to parse such files without any problems during tokenizing.
In this case, you can read the file as lines and separate it later using the delimiter and create a dataframe out of it. This is helpful when you have varying lengths of rows.
In the below example,
- the file is read as lines by specifying the separator as a new line using
sep='n'. Now the file will be tokenized on each new line, and a single column will be available in the dataframe. - You can split the lines using the separator or regex and create different columns out of it.
expand=Trueexpands the split string into multiple columns.
Use the below snippet to read the file as lines and separate it using the separator.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, sep='n')
df = df[0].str.split('s|s', expand=True)
dfThis is how you can read the file as lines and later separate it to avoid problems while parsing the lines with an inconsistent number of columns.
Conclusion
To summarize, you’ve learned the causes of the Python Pandas Error tokenizing data and the different methods to solve it in different scenarios.
Different Errors while tokenizing data are,
Error tokenizing data. C error: Buffer overflow caught - possible malformed input fileParserError: Expected n fields in line x, saw mError tokenizing data. c error out of memory
Also learned the different engines available in the read_csv() method to parse the CSV file and the advantages and disadvantages of it.
You’ve also learned when to use the different methods appropriately.
If you have any questions, comment below.
You May Also Like
- How to write pandas dataframe to CSV
- How To Read An Excel File In Pandas – With Examples
- How To Read Excel With Multiple Sheets In Pandas?
- How To Solve Truth Value Of A Series Is Ambiguous. Use A.Empty, A.Bool(), A.Item(), A.Any() Or A.All()?
- How to solve xlrd.biffh.XLRDError: Excel xlsx file; not supported Error?
Содержание
- How to Solve Error Tokenizing Data on read_csv in Pandas
- Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
- Step 2: Use correct separator to solve Pandas tokenizing error
- Step 3: Use different engine for read_csv()
- Step 4: Identify the headers to solve Error Tokenizing Data
- Step 5: Autodetect skiprows for read_csv()
- Conclusion
- How To Fix pandas.parser.CParserError: Error tokenizing data
- Understanding why the error is raised and how to deal with it when reading CSV files in pandas
- Introduction
- Reproducing the error
- Fixing the file manually
- Specifying line terminator
- Specifying the correct delimiter and headers
- Skipping rows
- Final Thoughts
- Python ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z (Example)
- Example Data & Software Libraries
- Reproduce the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
- Debug the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
- Video & Further Resources
- How to Solve Python Pandas Error Tokenizing Data Error?
- Cause of the Problem
- Finding the Problematic Line (Optional)
- Using Err_Bad_Lines Parameter
- Using Python Engine
- Using Proper Separator
- Using Line Terminator
- Using header=None
- Using Skiprows
- Reading As Lines and Separating
- Conclusion
How to Solve Error Tokenizing Data on read_csv in Pandas
In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
which we are going to read by — read_csv() method:
We will get an error:
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines=’skip’ in order to skip bad lines:
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
Note that delimiter is an alias for sep .
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
or skipping the last lines:
Note that we need to use python as the read_csv() engine for parameter — skipfooter .
In some cases header=None can help in order to solve the error:
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
Conclusion
In this post we saw how to investigate the error:
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
Источник
How To Fix pandas.parser.CParserError: Error tokenizing data
Understanding why the error is raised and how to deal with it when reading CSV files in pandas
Introduction
Importing data from csv files is probably the most commonly used way of instantiating pandas DataFrames. However, many times this could be a little bit tricky especially when the data included in the file are not in the expected form. In these cases, pandas parser may raise an error similar to the one reported below:
In today’s short guide we will discuss why this error is being raised in the first place and additionally, we will discuss a few ways that could eventually help you deal with it.
Reproducing the error
First, let’s try to reproduce the error using a small dataset I have prepared as part of this tutorial.
Now if we attempt to read in the file using read_csv :
we are going to get the following error
The error is pretty clear as it indicates that on the 4th line instead of 4, 6 fields were observed (and by the way, the same issue occurs in the last line as well).
By default, read_csv uses comma ( , ) as the delimiter but clearly, two lines in the file have more separators than expected. The expected number in this occasion is actually 4, since our headers (i.e. the first line of the file) contains 4 fields separated by commas.
Fixing the file manually
The most obvious solution to the problem, is to fix the data file manually by removing the extra separators in the lines causing us troubles. This is actually the best solution (assuming that you have specified the right delimiters, headers etc. when calling read_csv function). However, this may be quite tricky and painful when you need to deal with large files containing thousands of lines.
Specifying line terminator
Another cause of this error may be related to some carriage returns (i.e. ‘r’ ) in the data. In some occasions this is actually introduced by pandas.to_csv() method. When writing pandas DataFrames to CSV files, a carriage return is added to column names in which the method will then write the subsequent column names to the first column of the pandas DataFrame. And thus we’ll end up with different number of columns in the first rows.
If that’s the case, then you can explicitly specify the line terminator to ‘n’ using the corresponding parameter when calling read_csv() :
Specifying the correct delimiter and headers
The error may also be related to the delimiters and/or headers (not) specified when calling read_csv . Make sure to pass both the correct separator and headers.
For example, the arguments below specify that ; is the delimiter used to separate columns (by default commas are used as delimiters) and that the file does not contain any headers at all.
Skipping rows
Skipping rows that are causing the error should be your last resort and I would personally discourage you from doing so, but I guess there are certain use cases where this may be acceptable.
If that’s the case, then you can do so by setting error_bad_lines to False when calling read_csv function:
As you can see from the output above, the lines that they were causing errors were actually skipped and we can now move on with whatever we’d like to do with our pandas DataFrame.
Final Thoughts
In today’s short guide, we discussed a few cases where pandas.errors.ParserError: Error tokenizing data is raised by the pandas parser when reading csv files into pandas DataFrames.
Additionally, we showcased how to deal with the error by fixing the errors or typos in the data file itself, or by specifying the appropriate line terminator. Finally, we also discussed how to skip lines causing errors but keep in mind that in most of the cases this should be avoided.
Become a member and read every story on Medium. Your membership fee directly supports me and other writers you read. You’ll also get full access to every story on Medium.
Источник
Python ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z (Example)
In this tutorial you’ll learn how to fix the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language.
The article consists of the following information:
Let’s get started.
Example Data & Software Libraries
Consider the CSV file illustrated below as a basis for this tutorial:
You may already note that rows 4 and 6 contain one value too much. Those two rows contain four different values, but the other rows contain only three values.
Let’s assume that we want to read this CSV file as a pandas DataFrame into Python.
For this, we first have to import the pandas library:
import pandas as pd # Load pandas
Let’s move on to the examples!
Reproduce the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
In this section, I’ll show how to replicate the error message “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z”.
Let’s assume that we want to read our example CSV file using the default settings of the read_csv function. Then, we might try to import our data as shown below:
data_import = pd.read_csv(‘data.csv’) # Try to import CSV file # ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
Unfortunately, the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” is returned after executing the Python syntax above.
The reason for this is that our CSV file contains too many values in some of the rows.
In the next section, I’ll show an easy solution for this problem. So keep on reading…
Debug the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
In this example, I’ll explain an easy fix for the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language.
We can ignore all lines in our CSV file that are formatted wrongly by specifying the error_bad_lines argument to False.
Have a look at the example code below:
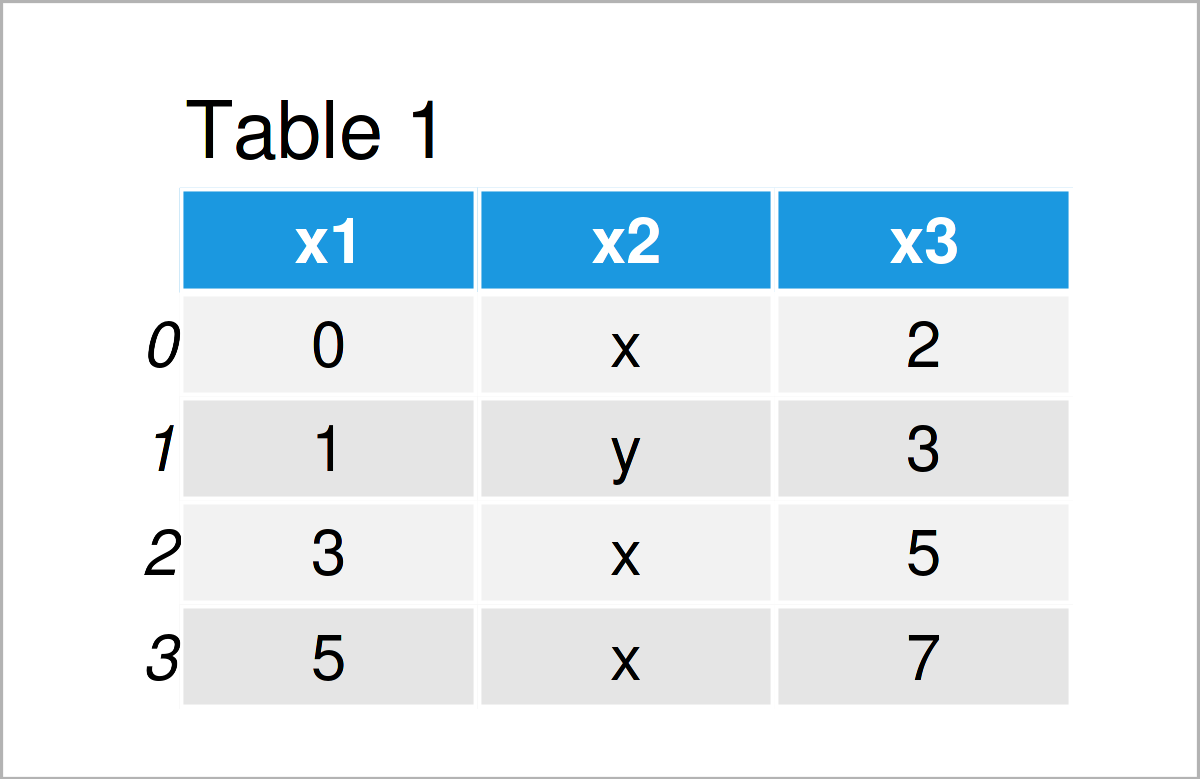
data_import = pd.read_csv(‘data.csv’, # Remove rows with errors error_bad_lines = False) print(data_import) # Print imported pandas DataFrame
As shown in Table 2, we have created a valid pandas DataFrame output using the previous code. As you can see, we have simply skipped the rows with too many values.
This is a simply trick that usually works. However, please note that this trick should be done with care, since the discussed error message typically points to more general issues with your data.
For that reason, it’s advisable to investigate why some of the rows are not formatted properly.
For this, I can also recommend this thread on Stack Overflow. It discusses how to identify wrong lines, and it also discusses other less common reasons for the error message “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z”.
Video & Further Resources
Have a look at the following video on my YouTube channel. In the video, I’m explaining the Python codes of this tutorial:
The YouTube video will be added soon.
Furthermore, you might read the other articles on this website. You can find some interesting tutorials below:
Summary: In this article, I have explained how to handle the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language. If you have any further questions or comments, let me know in the comments. Furthermore, don’t forget to subscribe to my email newsletter to get updates on new articles.
Источник
How to Solve Python Pandas Error Tokenizing Data Error?
While reading a CSV file, you may get the “Pandas Error Tokenizing Data“. This mostly occurs due to the incorrect data in the CSV file.
You can solve python pandas error tokenizing data error by ignoring the offending lines using error_bad_lines=False .
In this tutorial, you’ll learn the cause and how to solve the error tokenizing data error.
If you’re in Hurry
You can use the below code snippet to solve the tokenizing error. You can solve the error by ignoring the offending lines and suppressing errors.
Snippet
If You Want to Understand Details, Read on…
In this tutorial, you’ll learn the causes for the exception “Error Tokenizing Data” and how it can be solved.
Table of Contents
Cause of the Problem
- CSV file has two header lines
- Different separator is used
- r – is a new line character and it is present in column names which makes subsequent column names to be read as next line
- Lines of the CSV files have inconsistent number of columns
In the case of invalid rows which has an inconsistent number of columns, you’ll see an error as Expected 1 field in line 12, saw m . This means it expected only 1 field in the CSV file but it saw 12 values after tokenizing it. Hence, it doesn’t know how the tokenized values need to be handled. You can solve the errors by using one of the options below.
Finding the Problematic Line (Optional)
If you want to identify the line which is creating the problem while reading, you can use the below code snippet.
It uses the CSV reader. hence it is similar to the read_csv() method.
Snippet
Using Err_Bad_Lines Parameter
When there is insufficient data in any of the rows, the tokenizing error will occur.
You can skip such invalid rows by using the err_bad_line parameter within the read_csv() method.
This parameter controls what needs to be done when a bad line occurs in the file being read.
- False – Errors will be suppressed for Invalid lines
- True – Errors will be thrown for invalid lines
Use the below snippet to read the CSV file and ignore the invalid lines. Only a warning will be shown with the line number when there is an invalid lie found.
Snippet
In this case, the offending lines will be skipped and only the valid lines will be read from CSV and a dataframe will be created.
Using Python Engine
There are two engines supported in reading a CSV file. C engine and Python Engine.
C Engine
- Faster
- Uses C language to parse the CSV file
- Supports float_precision
- Cannot automatically detect the separator
- Doesn’t support skipping footer
Python Engine
- Slower when compared to C engine but its feature complete
- Uses Python language to parse the CSV file
- Doesn’t support float_precision . Not required with Python
- Can automatically detect the separator
- Supports skipping footer
Using the python engine can solve the problems faced while parsing the files.
For example, When you try to parse large CSV files, you may face the Error tokenizing data. c error out of memory. Using the python engine can solve the memory issues while parsing such big CSV files using the read_csv() method.
Use the below snippet to use the Python engine for reading the CSV file.
Snippet
This is how you can use the python engine to parse the CSV file.
Optionally, this could also solve the error Error tokenizing data. c error out of memory when parsing the big CSV files.
Using Proper Separator
CSV files can have different separators such as tab separator or any other special character such as ; . In this case, an error will be thrown when reading the CSV file, if the default C engine is used.
You can parse the file successfully by specifying the separator explicitly using the sep parameter.
As an alternative, you can also use the python engine which will automatically detect the separator and parse the file accordingly.
Snippet
This is how you can specify the separator explicitly which can solve the tokenizing errors while reading the CSV files.
Using Line Terminator
CSV file can contain r carriage return for separating the lines instead of the line separator n .
In this case, you’ll face CParserError: Error tokenizing data. C error: Buffer overflow caught — possible malformed input file when the line contains the r instead on n .
You can solve this error by using the line terminator explicitly using the lineterminator parameter.
Snippet
This is how you can use the line terminator to parse the files with the terminator r .
CSV files can have incomplete headers which can cause tokenizing errors while parsing the file.
You can use header=None to ignore the first line headers while reading the CSV files.
This will parse the CSV file without headers and create a data frame. You can also add headers to column names by adding columns attribute to the read_csv() method.
Snippet
This is how you can ignore the headers which are incomplete and cause problems while reading the file.
Using Skiprows
CSV files can have headers in more than one row. This can happen when data is grouped into different sections and each group is having a name and has columns in each section.
In this case, you can ignore such rows by using the skiprows parameter. You can pass the number of rows to be skipped and the data will be read after skipping those number of rows.
Use the below snippet to skip the first two rows while reading the CSV file.
Snippet
This is how you can skip or ignore the erroneous headers while reading the CSV file.
Reading As Lines and Separating
In a CSV file, you may have a different number of columns in each row. This can occur when some of the columns in the row are considered optional. You may need to parse such files without any problems during tokenizing.
In this case, you can read the file as lines and separate it later using the delimiter and create a dataframe out of it. This is helpful when you have varying lengths of rows.
In the below example,
- the file is read as lines by specifying the separator as a new line using sep=’n’ . Now the file will be tokenized on each new line, and a single column will be available in the dataframe.
- You can split the lines using the separator or regex and create different columns out of it.
- expand=True expands the split string into multiple columns.
Use the below snippet to read the file as lines and separate it using the separator.
Snippet
This is how you can read the file as lines and later separate it to avoid problems while parsing the lines with an inconsistent number of columns.
Conclusion
To summarize, you’ve learned the causes of the Python Pandas Error tokenizing data and the different methods to solve it in different scenarios.
Different Errors while tokenizing data are,
- Error tokenizing data. C error: Buffer overflow caught — possible malformed input file
- ParserError: Expected n fields in line x, saw m
- Error tokenizing data. c error out of memory
Also learned the different engines available in the read_csv() method to parse the CSV file and the advantages and disadvantages of it.
You’ve also learned when to use the different methods appropriately.
Источник