Recently, I burned about 3 hours trying to load a large CSV file into Python Pandas using the read_csv function, only to consistently run into the following error:
ParserError Traceback (most recent call last) <ipython-input-6-b51ad8562823> in <module>() ... pandas_libsparsers.pyx in pandas._libs.parsers.TextReader.read (pandas_libsparsers.c:10862)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_low_memory (pandas_libsparsers.c:11138)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_rows (pandas_libsparsers.c:11884)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._tokenize_rows (pandas_libsparsers.c:11755)() pandas_libsparsers.pyx in pandas._libs.parsers.raise_parser_error (pandas_libsparsers.c:28765)() Error tokenizing data. C error: EOF inside string starting at line XXXX
“Error tokenising data. C error: EOF inside string starting at line”.
There was an erroneous character about 5000 lines into the CSV file that prevented the Pandas CSV parser from reading the entire file. Excel had no problems opening the file, and no amount of saving/re-saving/changing encodings was working. Manually removing the offending line worked, but ultimately, another character 6000 lines further into the file caused the same issue.
The solution was to use the parameter engine=’python’ in the read_csv function call. The Pandas CSV parser can use two different “engines” to parse CSV file – Python or C (default).
pandas.read_csv(filepath_or_buffer, sep=', ', delimiter=None,
header='infer', names=None,
index_col=None, usecols=None, squeeze=False,
..., engine=None, ...)
engine : {‘c’, ‘python’}, optional
Parser engine to use. The C engine is faster while the python engine is currently more feature-complete.
The Python engine is described as “slower, but more feature complete” in the Pandas documentation. However, in this case, the python engine sorts the problem, without a massive time impact (overall file size was approx 200,000 rows).
UnicodeDecodeError
The other big problem with reading in CSV files that I come across is the error:
“UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
Character encoding issues can be usually fixed by opening the CSV file in Sublime Text, and then “Save with encoding” choosing UTF-8. Then adding the encoding=’utf8′ to the pandas.read_csv command allows Pandas to open the CSV file without trouble. This error appears regularly for me with files saved in Excel.
encoding : str, default None
Содержание
- Pandas CSV error: Error tokenizing data. C error: EOF inside string starting at line
- Tokenizing Error
- “Error tokenising data. C error: EOF inside string starting at line”.
- UnicodeDecodeError
- “UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
- pandas.errors.ParserError: Error tokenizing data. C error: EOF inside string starting at line x #22140
- Comments
- Error
- Problem description
- Expected Output
- Output of pd.show_versions()
- [paste the output of pd.show_versions() here below this line] INSTALLED VERSIONS
- read_csv C-engine CParserError: Error tokenizing data #11166
- Comments
- How To Fix pandas.parser.CParserError: Error tokenizing data
- Understanding why the error is raised and how to deal with it when reading CSV files in pandas
- Introduction
- Reproducing the error
- Fixing the file manually
- Specifying line terminator
- Specifying the correct delimiter and headers
- Skipping rows
- Final Thoughts
- How To Fix pandas.parser.CParserError: Error tokenizing data
- Understanding why the error is raised and how to deal with it when reading CSV files in pandas
- Introduction
- Reproducing the error
- Fixing the file manually
- Specifying line terminator
- Specifying the correct delimiter and headers
- Skipping rows
- Final Thoughts
Pandas CSV error: Error tokenizing data. C error: EOF inside string starting at line
Tokenizing Error
Recently, I burned about 3 hours trying to load a large CSV file into Python Pandas using the read_csv function, only to consistently run into the following error:
“Error tokenising data. C error: EOF inside string starting at line”.
There was an erroneous character about 5000 lines into the CSV file that prevented the Pandas CSV parser from reading the entire file. Excel had no problems opening the file, and no amount of saving/re-saving/changing encodings was working. Manually removing the offending line worked, but ultimately, another character 6000 lines further into the file caused the same issue.
The solution was to use the parameter engine=’python’ in the read_csv function call. The Pandas CSV parser can use two different “engines” to parse CSV file – Python or C (default).
Parser engine to use. The C engine is faster while the python engine is currently more feature-complete.
The Python engine is described as “slower, but more feature complete” in the Pandas documentation. However, in this case, the python engine sorts the problem, without a massive time impact (overall file size was approx 200,000 rows).
UnicodeDecodeError
The other big problem with reading in CSV files that I come across is the error:
“UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
Character encoding issues can be usually fixed by opening the CSV file in Sublime Text, and then “Save with encoding” choosing UTF-8. Then adding the encoding=’utf8′ to the pandas.read_csv command allows Pandas to open the CSV file without trouble. This error appears regularly for me with files saved in Excel.
encoding : str, default None
Источник
pandas.errors.ParserError: Error tokenizing data. C error: EOF inside string starting at line x #22140
Error
Problem description
The current error handling of error while parsing double quote («) in ‘C’ engine is vague. If the data is sensitive there is very less chance that you will get to have a look at the actual data. So, if the error is more specific it would be awesome. In my company we get less data for development which is masked of`course, but in production it is unclear why and where errors occurred.
If not as an error, display a warning saying that a » is present in between a line.
Expected Output
There is a weird character («) leading to EOF. » is taken as end of string.
Output of pd.show_versions()
[paste the output of pd.show_versions() here below this line]
INSTALLED VERSIONS
commit: None
python: 3.7.0.final.0
python-bits: 32
OS: Windows
OS-release: 10
machine: AMD64
processor: Intel64 Family 6 Model 158 Stepping 10, GenuineIntel
byteorder: little
LC_ALL: None
LANG: None
LOCALE: None.None
pandas: 0.23.3
pytest: None
pip: 18.0
setuptools: 40.0.0
Cython: None
numpy: 1.15.0
scipy: None
pyarrow: None
xarray: None
IPython: None
sphinx: None
patsy: None
dateutil: 2.7.3
pytz: 2018.5
blosc: None
bottleneck: None
tables: None
numexpr: None
feather: None
matplotlib: None
openpyxl: None
xlrd: None
xlwt: None
xlsxwriter: None
lxml: None
bs4: None
html5lib: None
sqlalchemy: None
pymysql: None
psycopg2: None
jinja2: None
s3fs: None
fastparquet: None
pandas_gbq: None
pandas_datareader: None
The text was updated successfully, but these errors were encountered:
Источник
read_csv C-engine CParserError: Error tokenizing data #11166
I have encountered a dataset where the C-engine read_csv has problems. I am unsure of the exact issue but I have narrowed it down to a single row which I have pickled and uploaded it to dropbox. If you obtain the pickle try the following:
I get the following exception:
If you try and read the CSV using the python engine then no exception is thrown:
Suggesting that the issue is with read_csv and not to_csv. The versions I using are:
The text was updated successfully, but these errors were encountered:
Your second-to-last line includes an ‘r’ break. I think it’s a bug, but one workaround is to open in universal-new-line mode.
I’m encountering this error as well. Using the method suggested by @chris-b1 causes the following error:
I have also found this issue when reading a large csv file with the default egine. If I use engine=’python’ then it works fine.
I missed @alfonsomhc answer because it just looked like a comment.
had the same issue trying to read a folder not a csv file
Has anyone investigated this issue? It’s killing performance when using read_csv in a keras generator.
The original data provided is no longer available so the issue is not reproducible. Closing as it’s not clear what the issue is, but @dgrahn or anyone else if you can provide a reproducible example we can reopen
@WillAyd Let me know if you need additional info.
Since GitHub doesn’t accept CSVs, I changed the extension to .txt.
Here’s the code which will trigger the exception.
Here’s the exception from Windows 10, using Anaconda.
And the same on RedHat.
@dgrahn I have downloaded debug.txt and I get the following if you run pd.read_csv(‘debug.xt’, header=None) on a mac:
ParserError: Error tokenizing data. C error: Expected 204 fields in line 3, saw 2504
Which is different from the Buffer overflow caught error originally described.
I have inspected the debug.txt file and the first two lines have 204 columns but the 3rd line has 2504 columns. This would make the file unparsable and explains why an error is thrown.
Is this expected? GitHub could be doing some implicit conversion in the background between newline types («rn» and «n») that is messing up the uploaded example.
@joshlk Did you use the names=range(2504) option as described in the comment above?
Ok can now reproduce the error with pandas.read_csv(‘debug.csv’, chunksize=1000, names=range(2504)) .
It’s good to note that pandas.read_csv(‘debug.csv’, names=range(2504)) works fine and so its then unlikely related to the original bug but it is producing the same symptom.
@joshlk I could open a separate issue if that would be preferred.
pd.read_csv(open(‘test.csv’,’rU’), encoding=’utf-8′, engine=’python’)
Solved my problem.
I tried this approach and was able to upload large data files. But when I checked the dimension of the dataframe I saw that the number of rows have increased. What can be the logical regions for that?
@dheeman00 : I am facing the same problem as you with changing sizes. I have a dataframe of shape (100K, 21) and after using engine = ‘python’, it gives me a dataframe of shape (100,034,21) (without enging=’python’, I get the same error as OP). After comparing them, I figured the problem is with one of my columns that contains text field, some with unknown chars, and some of them are broken into two different rows (the second row with the continuation of the text has all other columns set to «nan»).
If you know your data well, playing with delimiters and maybe running a data cleaning before saving as CSV would be helpful. In my case, the data was too messy and too big (it was the subset of a bigger csv file) so I changed to Spark for data cleaning.
Источник
How To Fix pandas.parser.CParserError: Error tokenizing data
Understanding why the error is raised and how to deal with it when reading CSV files in pandas
Introduction
Importing data from csv files is probably the most commonly used way of instantiating pandas DataFrames. However, many times this could be a little bit tricky especially when the data included in the file are not in the expected form. In these cases, pandas parser may raise an error similar to the one reported below:
In today’s short guide we will discuss why this error is being raised in the first place and additionally, we will discuss a few ways that could eventually help you deal with it.
Reproducing the error
First, let’s try to reproduce the error using a small dataset I have prepared as part of this tutorial.
Now if we attempt to read in the file using read_csv :
we are going to get the following error
The error is pretty clear as it indicates that on the 4th line instead of 4, 6 fields were observed (and by the way, the same issue occurs in the last line as well).
By default, read_csv uses comma ( , ) as the delimiter but clearly, two lines in the file have more separators than expected. The expected number in this occasion is actually 4, since our headers (i.e. the first line of the file) contains 4 fields separated by commas.
Fixing the file manually
The most obvious solution to the problem, is to fix the data file manually by removing the extra separators in the lines causing us troubles. This is actually the best solution (assuming that you have specified the right delimiters, headers etc. when calling read_csv function). However, this may be quite tricky and painful when you need to deal with large files containing thousands of lines.
Specifying line terminator
Another cause of this error may be related to some carriage returns (i.e. ‘r’ ) in the data. In some occasions this is actually introduced by pandas.to_csv() method. When writing pandas DataFrames to CSV files, a carriage return is added to column names in which the method will then write the subsequent column names to the first column of the pandas DataFrame. And thus we’ll end up with different number of columns in the first rows.
If that’s the case, then you can explicitly specify the line terminator to ‘n’ using the corresponding parameter when calling read_csv() :
Specifying the correct delimiter and headers
The error may also be related to the delimiters and/or headers (not) specified when calling read_csv . Make sure to pass both the correct separator and headers.
For example, the arguments below specify that ; is the delimiter used to separate columns (by default commas are used as delimiters) and that the file does not contain any headers at all.
Skipping rows
Skipping rows that are causing the error should be your last resort and I would personally discourage you from doing so, but I guess there are certain use cases where this may be acceptable.
If that’s the case, then you can do so by setting error_bad_lines to False when calling read_csv function:
As you can see from the output above, the lines that they were causing errors were actually skipped and we can now move on with whatever we’d like to do with our pandas DataFrame.
Final Thoughts
In today’s short guide, we discussed a few cases where pandas.errors.ParserError: Error tokenizing data is raised by the pandas parser when reading csv files into pandas DataFrames.
Additionally, we showcased how to deal with the error by fixing the errors or typos in the data file itself, or by specifying the appropriate line terminator. Finally, we also discussed how to skip lines causing errors but keep in mind that in most of the cases this should be avoided.
Become a member and read every story on Medium. Your membership fee directly supports me and other writers you read. You’ll also get full access to every story on Medium.
Источник
How To Fix pandas.parser.CParserError: Error tokenizing data
Understanding why the error is raised and how to deal with it when reading CSV files in pandas
Introduction
Importing data from csv files is probably the most commonly used way of instantiating pandas DataFrames. However, many times this could be a little bit tricky especially when the data included in the file are not in the expected form. In these cases, pandas parser may raise an error similar to the one reported below:
In today’s short guide we will discuss why this error is being raised in the first place and additionally, we will discuss a few ways that could eventually help you deal with it.
Reproducing the error
First, let’s try to reproduce the error using a small dataset I have prepared as part of this tutorial.
Now if we attempt to read in the file using read_csv :
we are going to get the following error
The error is pretty clear as it indicates that on the 4th line instead of 4, 6 fields were observed (and by the way, the same issue occurs in the last line as well).
By default, read_csv uses comma ( , ) as the delimiter but clearly, two lines in the file have more separators than expected. The expected number in this occasion is actually 4, since our headers (i.e. the first line of the file) contains 4 fields separated by commas.
Fixing the file manually
The most obvious solution to the problem, is to fix the data file manually by removing the extra separators in the lines causing us troubles. This is actually the best solution (assuming that you have specified the right delimiters, headers etc. when calling read_csv function). However, this may be quite tricky and painful when you need to deal with large files containing thousands of lines.
Specifying line terminator
Another cause of this error may be related to some carriage returns (i.e. ‘r’ ) in the data. In some occasions this is actually introduced by pandas.to_csv() method. When writing pandas DataFrames to CSV files, a carriage return is added to column names in which the method will then write the subsequent column names to the first column of the pandas DataFrame. And thus we’ll end up with different number of columns in the first rows.
If that’s the case, then you can explicitly specify the line terminator to ‘n’ using the corresponding parameter when calling read_csv() :
Specifying the correct delimiter and headers
The error may also be related to the delimiters and/or headers (not) specified when calling read_csv . Make sure to pass both the correct separator and headers.
For example, the arguments below specify that ; is the delimiter used to separate columns (by default commas are used as delimiters) and that the file does not contain any headers at all.
Skipping rows
Skipping rows that are causing the error should be your last resort and I would personally discourage you from doing so, but I guess there are certain use cases where this may be acceptable.
If that’s the case, then you can do so by setting error_bad_lines to False when calling read_csv function:
As you can see from the output above, the lines that they were causing errors were actually skipped and we can now move on with whatever we’d like to do with our pandas DataFrame.
Final Thoughts
In today’s short guide, we discussed a few cases where pandas.errors.ParserError: Error tokenizing data is raised by the pandas parser when reading csv files into pandas DataFrames.
Additionally, we showcased how to deal with the error by fixing the errors or typos in the data file itself, or by specifying the appropriate line terminator. Finally, we also discussed how to skip lines causing errors but keep in mind that in most of the cases this should be avoided.
Become a member and read every story on Medium. Your membership fee directly supports me and other writers you read. You’ll also get full access to every story on Medium.
Источник
Data
Col1|Col2|Col3|Col4 Str1|1|p|num1 Str2|2|q|num2 Str3|3|"|num3 Str4|4|s|num4
Error
>>> import pandas as pd >>> df = pd.read_csv('./sample.txt', sep='|') Traceback (most recent call last): File "<stdin>", line 1, in <module> File ".envlibsite-packagespandasioparsers.py", line 678, in parser_f return _read(filepath_or_buffer, kwds) File ".envlibsite-packagespandasioparsers.py", line 446, in _read data = parser.read(nrows) File ".envlibsite-packagespandasioparsers.py", line 1036, in read ret = self._engine.read(nrows) File ".envlibsite-packagespandasioparsers.py", line 1848, in read data = self._reader.read(nrows) File "pandas_libsparsers.pyx", line 876, in pandas._libs.parsers.TextReader.read File "pandas_libsparsers.pyx", line 891, in pandas._libs.parsers.TextReader._read_low_memory File "pandas_libsparsers.pyx", line 945, in pandas._libs.parsers.TextReader._read_rows File "pandas_libsparsers.pyx", line 932, in pandas._libs.parsers.TextReader._tokenize_rows File "pandas_libsparsers.pyx", line 2112, in pandas._libs.parsers.raise_parser_error pandas.errors.ParserError: Error tokenizing data. C error: EOF inside string starting at line 3 >>>
Problem description
The current error handling of error while parsing double quote («) in ‘C’ engine is vague. If the data is sensitive there is very less chance that you will get to have a look at the actual data. So, if the error is more specific it would be awesome. In my company we get less data for development which is masked of`course, but in production it is unclear why and where errors occurred.
If not as an error, display a warning saying that a » is present in between a line.
Expected Output
There is a weird character («) leading to EOF. » is taken as end of string.
Output of pd.show_versions()
[paste the output of pd.show_versions() here below this line]
INSTALLED VERSIONS
commit: None
python: 3.7.0.final.0
python-bits: 32
OS: Windows
OS-release: 10
machine: AMD64
processor: Intel64 Family 6 Model 158 Stepping 10, GenuineIntel
byteorder: little
LC_ALL: None
LANG: None
LOCALE: None.None
pandas: 0.23.3
pytest: None
pip: 18.0
setuptools: 40.0.0
Cython: None
numpy: 1.15.0
scipy: None
pyarrow: None
xarray: None
IPython: None
sphinx: None
patsy: None
dateutil: 2.7.3
pytz: 2018.5
blosc: None
bottleneck: None
tables: None
numexpr: None
feather: None
matplotlib: None
openpyxl: None
xlrd: None
xlwt: None
xlsxwriter: None
lxml: None
bs4: None
html5lib: None
sqlalchemy: None
pymysql: None
psycopg2: None
jinja2: None
s3fs: None
fastparquet: None
pandas_gbq: None
pandas_datareader: None
Pandas CSV error: Error tokenizing data. C error: EOF inside string starting at line
Tokenizing Error
Recently, I burned about 3 hours trying to load a large CSV file into Python Pandas using the read_csv function, only to consistently run into the following error:
“Error tokenising data. C error: EOF inside string starting at line”.
There was an erroneous character about 5000 lines into the CSV file that prevented the Pandas CSV parser from reading the entire file. Excel had no problems opening the file, and no amount of saving/re-saving/changing encodings was working. Manually removing the offending line worked, but ultimately, another character 6000 lines further into the file caused the same issue.
The solution was to use the parameter engine=’python’ in the read_csv function call. The Pandas CSV parser can use two different “engines” to parse CSV file – Python or C (default).
Parser engine to use. The C engine is faster while the python engine is currently more feature-complete.
The Python engine is described as “slower, but more feature complete” in the Pandas documentation. However, in this case, the python engine sorts the problem, without a massive time impact (overall file size was approx 200,000 rows).
UnicodeDecodeError
The other big problem with reading in CSV files that I come across is the error:
“UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
Character encoding issues can be usually fixed by opening the CSV file in Sublime Text, and then “Save with encoding” choosing UTF-8. Then adding the encoding=’utf8′ to the pandas.read_csv command allows Pandas to open the CSV file without trouble. This error appears regularly for me with files saved in Excel.
encoding : str, default None
Источник
pandas.errors.ParserError: Error tokenizing data. C error: EOF inside string starting at line x #22140
Comments
Error
Problem description
The current error handling of error while parsing double quote («) in ‘C’ engine is vague. If the data is sensitive there is very less chance that you will get to have a look at the actual data. So, if the error is more specific it would be awesome. In my company we get less data for development which is masked of`course, but in production it is unclear why and where errors occurred.
If not as an error, display a warning saying that a » is present in between a line.
Expected Output
There is a weird character («) leading to EOF. » is taken as end of string.
Output of pd.show_versions()
[paste the output of pd.show_versions() here below this line]
INSTALLED VERSIONS
commit: None
python: 3.7.0.final.0
python-bits: 32
OS: Windows
OS-release: 10
machine: AMD64
processor: Intel64 Family 6 Model 158 Stepping 10, GenuineIntel
byteorder: little
LC_ALL: None
LANG: None
LOCALE: None.None
pandas: 0.23.3
pytest: None
pip: 18.0
setuptools: 40.0.0
Cython: None
numpy: 1.15.0
scipy: None
pyarrow: None
xarray: None
IPython: None
sphinx: None
patsy: None
dateutil: 2.7.3
pytz: 2018.5
blosc: None
bottleneck: None
tables: None
numexpr: None
feather: None
matplotlib: None
openpyxl: None
xlrd: None
xlwt: None
xlsxwriter: None
lxml: None
bs4: None
html5lib: None
sqlalchemy: None
pymysql: None
psycopg2: None
jinja2: None
s3fs: None
fastparquet: None
pandas_gbq: None
pandas_datareader: None
The text was updated successfully, but these errors were encountered:
Источник
How To Fix pandas.parser.CParserError: Error tokenizing data
Understanding why the error is raised and how to deal with it when reading CSV files in pandas
Introduction
Importing data from csv files is probably the most commonly used way of instantiating pandas DataFrames. However, many times this could be a little bit tricky especially when the data included in the file are not in the expected form. In these cases, pandas parser may raise an error similar to the one reported below:
In today’s short guide we will discuss why this error is being raised in the first place and additionally, we will discuss a few ways that could eventually help you deal with it.
Reproducing the error
First, let’s try to reproduce the error using a small dataset I have prepared as part of this tutorial.
Now if we attempt to read in the file using read_csv :
we are going to get the following error
The error is pretty clear as it indicates that on the 4th line instead of 4, 6 fields were observed (and by the way, the same issue occurs in the last line as well).
By default, read_csv uses comma ( , ) as the delimiter but clearly, two lines in the file have more separators than expected. The expected number in this occasion is actually 4, since our headers (i.e. the first line of the file) contains 4 fields separated by commas.
Fixing the file manually
The most obvious solution to the problem, is to fix the data file manually by removing the extra separators in the lines causing us troubles. This is actually the best solution (assuming that you have specified the right delimiters, headers etc. when calling read_csv function). However, this may be quite tricky and painful when you need to deal with large files containing thousands of lines.
Specifying line terminator
Another cause of this error may be related to some carriage returns (i.e. ‘r’ ) in the data. In some occasions this is actually introduced by pandas.to_csv() method. When writing pandas DataFrames to CSV files, a carriage return is added to column names in which the method will then write the subsequent column names to the first column of the pandas DataFrame. And thus we’ll end up with different number of columns in the first rows.
If that’s the case, then you can explicitly specify the line terminator to ‘n’ using the corresponding parameter when calling read_csv() :
Specifying the correct delimiter and headers
The error may also be related to the delimiters and/or headers (not) specified when calling read_csv . Make sure to pass both the correct separator and headers.
For example, the arguments below specify that ; is the delimiter used to separate columns (by default commas are used as delimiters) and that the file does not contain any headers at all.
Skipping rows
Skipping rows that are causing the error should be your last resort and I would personally discourage you from doing so, but I guess there are certain use cases where this may be acceptable.
If that’s the case, then you can do so by setting error_bad_lines to False when calling read_csv function:
As you can see from the output above, the lines that they were causing errors were actually skipped and we can now move on with whatever we’d like to do with our pandas DataFrame.
Final Thoughts
In today’s short guide, we discussed a few cases where pandas.errors.ParserError: Error tokenizing data is raised by the pandas parser when reading csv files into pandas DataFrames.
Additionally, we showcased how to deal with the error by fixing the errors or typos in the data file itself, or by specifying the appropriate line terminator. Finally, we also discussed how to skip lines causing errors but keep in mind that in most of the cases this should be avoided.
Become a member and read every story on Medium. Your membership fee directly supports me and other writers you read. You’ll also get full access to every story on Medium.
Источник
How to Solve Error Tokenizing Data on read_csv in Pandas
In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
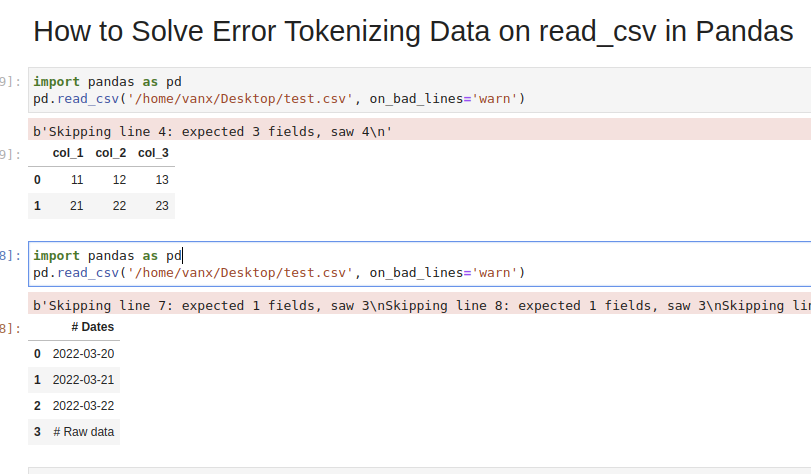
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
which we are going to read by — read_csv() method:
We will get an error:
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines=’skip’ in order to skip bad lines:
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
Note that delimiter is an alias for sep .
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
Step 4: Identify the headers to solve Error Tokenizing Data
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
or skipping the last lines:
Note that we need to use python as the read_csv() engine for parameter — skipfooter .
In some cases header=None can help in order to solve the error:
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
Conclusion
In this post we saw how to investigate the error:
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
Источник
read_csv() & EOF character in string cause parsing issue #5500
Comments
While importing large text files using read_csv we occasionally get an EOF (End of File ) character within a string, which causes an exception: «Error tokenizing data. C error: EOF inside string starting at line. 844863» . This occurs even with «error_bad_lines = False»..
Further, the line stated in the error message is not the line containing the EOF character. In this particular case the actual row was approx. 230 rows before the one stated, which hinders exception handling. (I now see this difference was caused by other «bad_lines» that were being skipped — the quoted error line is correct but the imported rows was less.)
I feel it would be appropriate if «error_bad_lines = False» handled this exception and allowed such rows to be skipped.
I note that when importing this text file into Excel, the «premature» EOF is simply ignored.
We are running on Windows 8 , with python version 2.7 and pandas version 0.12
The text was updated successfully, but these errors were encountered:
Further investigation using a hex editor has revealed what is going on:
- I added 0x1A («EOF») to a different file and it did not cause any problems. Pandas read_csv imported it without error.
- I parsed every line of the problematic CSV individually, until I isolated the one causing the problem. It was over 3000 rows after the stated row number in the error message.
- the row in question had a column with a double quote mark following the delimiter — there were not supposed to be any quote marks in the file. There was no second double quote in the column, or on the row
- I think the quote mark caused the import to look for a second terminating double quote, ignoring column delimiters and end of line markers until it reached the end of the file. When it didn’t find one before the end of the file, Im speculating that triggered the «EOF inside a string» error message.
- I was able to work around the problem by setting the quotechar to be the same as the delimiter, while tells read_csv to ignore all quotes. It now imports the file perfectly.
- I still think «error_bad_lines» should catch this by checking if any row contains a column with a missing terminating quote.
- one reason I think this is important is that by adding a second such double quote, many lines apart, I was able to «fool» the system into skipping all the intervening lines, even though only two rows had an error.
@stephenjshaw are you able to try this on the master branch?
On pandas 0.13.1, I had the exact same problem and solution.
I am having the same issue and cannot find any offending characters in the lines near the line number given. Is there some way to search for weird characters given I have no clue where the issue is?
Источник
Solution:
Add when reading files quoting=csv.QUOTE_ NONE
data = pd.read_ csv(path + ‘/’ + fn,quoting=csv.QUOTE_ NONE)
Quote mode is no reference. When reading, it is considered that the content is not surrounded by the default reference character (“).
Relevant knowledge points:
pandas.read_ CSV parameters
quoting : int or csv.QUOTE_* instance, default 0
Controls quotation mark constants in CSV.
Optional quote_ MINIMAL (0), QUOTE_ ALL (1), QUOTE_ NONNUMERIC (2) ,QUOTE_ NONE (3)
Other similar errors
1、pandas.errors.ParserError: Error tokenizing data. C error: Expected * fields in line *, saw *
solve:
Method A. add parameters when reading files error_ bad_ Lines = false # add parameters
data= pd.read_ csv(data_ file, error_ bad_ lines=False)
When reading CSV files, the separator defaults to comma. Analysis shows that a cell in the read data contains two fields, that is, the value may contain two commas
Method B. open the file to another format required by the dataset. Do not be lazy and modify the suffix directly. For example, some formats can be modified by converting Excel to CSV and saving manually to ensure uniform format
The fundamental reason is that the data format is incorrect, which makes it impossible to read correctly. We should solve it from the aspect of file content format
Read More:
- Python error: pandas.errors.ParserError: Error tokenizing data. C error: Expected 3……
- Solution pandas.errors.ParserError : Error tokenizing data. C error: Buffer overflow caught
- How to Fix Parser rerror: error tokenizing data. C error: expected 2 fields in line 53, saw 3
- C language string processing error warning, c4996, sprintf, predicted, c4996, strcpy, c4996, strcat
- In scan(file = file, what = what, sep = sep, quote = quote, dec = dec, : EOF within quoted string
- How to Fix Sklearn ValueError: This solver needs samples of at least 2 classes in the data, but the data
- Differences between length() size() and C strlen() of C + + string member functions
- Error 1136 (21s01): column count doesn’t match value count at row 1
- Solve the problem of error: cannot pass objects of non trivially copyable type ‘STD:: String’ in C / C + +
- Error c2951: template declaration can only be used at the global, namespace, or class scope. Error c2598: link specification must be at the global scope
- MySQL error code 1217 (ER_ROW_IS_REFERENCED): Cannot delete or update a parent row: a foreign key co
- Python conversion hex to string, high and low data processing
- Freemarker Failed at: ${ findObj.applyDateStart? string(“yyyy-MM… Template
- SQL Error (3621): String or binary data would be truncated The statement has been terminated. */
- String operation to delete the character at the specified position
- Split keyword in ABAP when the separator is at the beginning and end of the string
- C++ string substr()
- A repeated string is composed of two identical strings. For example, abcabc is a repeated string with length of 6, while abcba does not have a duplicate string. Given any string, please help Xiaoqiang find the longest repeated substring.
- Under Ubuntu environment, eclipse writes C program, and error starting process is reported
- C # generate data in DataGridView into XML file
This article is originally published at https://www.shanelynn.ie
Tokenizing Error
Recently, I burned about 3 hours trying to load a large CSV file into Python Pandas using the read_csv function, only to consistently run into the following error:
ParserError Traceback (most recent call last) <ipython-input-6-b51ad8562823> in <module>() ... pandas_libsparsers.pyx in pandas._libs.parsers.TextReader.read (pandas_libsparsers.c:10862)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_low_memory (pandas_libsparsers.c:11138)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_rows (pandas_libsparsers.c:11884)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._tokenize_rows (pandas_libsparsers.c:11755)() pandas_libsparsers.pyx in pandas._libs.parsers.raise_parser_error (pandas_libsparsers.c:28765)() Error tokenizing data. C error: EOF inside string starting at line XXXX
“Error tokenising data. C error: EOF inside string starting at line”.
There was an erroneous character about 5000 lines into the CSV file that prevented the Pandas CSV parser from reading the entire file. Excel had no problems opening the file, and no amount of saving/re-saving/changing encodings was working. Manually removing the offending line worked, but ultimately, another character 6000 lines further into the file caused the same issue.
The solution was to use the parameter engine=’python’ in the read_csv function call. The Pandas CSV parser can use two different “engines” to parse CSV file – Python or C (default).
pandas.read_csv(filepath_or_buffer, sep=', ', delimiter=None,
header='infer', names=None,
index_col=None, usecols=None, squeeze=False,
..., engine=None, ...)
engine : {‘c’, ‘python’}, optional
Parser engine to use. The C engine is faster while the python engine is currently more feature-complete.
The Python engine is described as “slower, but more feature complete” in the Pandas documentation. However, in this case, the python engine sorts the problem, without a massive time impact (overall file size was approx 200,000 rows).
UnicodeDecodeError
The other big problem with reading in CSV files that I come across is the error:
“UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
Character encoding issues can be usually fixed by opening the CSV file in Sublime Text, and then “Save with encoding” choosing UTF-8. Then adding the encoding=’utf8′ to the pandas.read_csv command allows Pandas to open the CSV file without trouble. This error appears regularly for me with files saved in Excel.
encoding : str, default None
Thanks for visiting r-craft.org
This article is originally published at https://www.shanelynn.ie
Please visit source website for post related comments.
This week I posted a bit different post than my previous posts. Originally I was writing this in stackoverflow.com. However, before I posted my question, I made sure that I have already tried every options that might solve my issue. And it turned out that my last attempt worked! So I declined to post this in stackoverflow.com. Nevertheless, since I thought that I might need to re-look at this workaround in solving the CParseError in Pandas, thus, I decided to post it in my blog.
I would like to read a csv file using Python 3.5 and Pandas 0.18.1.
import pandas as pd
df = pd.read_csv(directory, skiprows=3)
However, I got the following error.
CParserError: Error tokenizing data. C error: EOF inside string starting at line 353996
If I check the csv file in the corresponding line, I found that this line is missing a double quotation mark.
with open(directory) as f:
content = f.readlines()
f.close()
for line in content:
print(line)
Here is the problematic line.
320597,83222081,Ratna,2031,320597073,NEKSA (TO),O-Pre Sales,Active,30136,8242015,RAYON ZEUS 8,20170222,"""Jl.Kamboja No.29, Lrg.Perburuan"", Lr.Perburuan, 1671040010, 16
So after ’16’ it should have been ended by , ". Here is the example of the other line that did not result in CParseError.
320597,83222080,Kartika,2031,320597073,NEKSA (TO),O-Pre Sales,Active,30136,8242015,RAYON ZEUS 8,20170222,"""Jl.Dwikora 2 No.2924, Lrg.Tinta Mas"", Lr.Tinta Mas, 1671040010, 1671040, 1671,16, "
In order to fix this issue, we have to do workaround. Currently what I am thinking is that modify the problematic line by manually assign the missing characters. However, if you have a better solution, please do let me and my readers know.
First, we have to know the line number of the problematic line. In order to get the line number, we need to catch the exception. Afterwards, I fed the exception message (after converting it to str) to the new method called fix_and_read_csv.
try:
df = pd.read_csv(directory)
except Exception as e:
e = str(e)
df = fix_and_read_csv(e, directory, append_chars=', "')
So what is this fix_and_read_csv? Basically, this method will:
- open the same
.csvfile that we have already opened before and returned an error - save it to temporary file read/write
- open in write mode the same
.csvfile - write the
.csvfile from the beginning - fix the problematic line by appending at the end of the line with the missing characters
- close and save the
.csvfile.
Here is my fix_and_read_csv code.
def fix_and_read_csv(exception_msg, directory, append_chars):
# step I: get the line number that is problematic
temp = exception_msg.split()
for item in temp:
try:
error_line = int(item)
except:
continue
# step II: fix the problematic line
modify_file(directory, error_line, append_chars)
# step III: create the dataframe and return it
df = pd.read_csv(directory, usecols=usecols, skiprows=skiprows)
return df
Inside the fix_and_read_csv method, I embedded one more method to read and then write back the fixed line to the same file. I named it modify_file. Here is the modify_file code.
def modify_file(filename, error_line_at, append_chars):
import tempfile
error_line_at = error_line_at + 6
#Create temporary file read/write
t = tempfile.NamedTemporaryFile(mode="r+")
#Open input file read-only
i = open(filename, 'r')
#Copy input file to temporary file, modifying as we go
for line in i:
t.write(line)
i.close() #Close input file
t.seek(0) #Rewind temporary file to beginning
o = open(filename, "w") #Reopen input file writable
#Overwriting original file with temporary file contents
i = -1
for line in t:
i += 1
if i != error_line_at: o.write(line)
else: o.write(line+append_chars)
t.close() #Close temporary file, will cause it to be deleted
Again, I am sorry I could not continue my previous post this time. The style of the post is also a bit different, because I just copied and pasted my post in stackoverflow.com to here. Nonetheless, I hope that you still enjoy reading this week’s post. As always, thanks for reading. Should there be any query or comment, please leave them down in the comment section below. See you guys in the next post!