In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
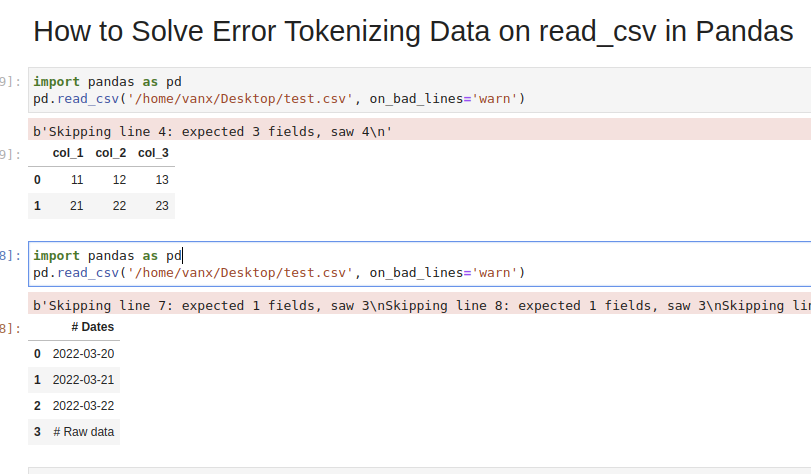
col_1,col_2,col_3
11,12,13
21,22,23
31,32,33,44
which we are going to read by — read_csv() method:
import pandas as pd
pd.read_csv('test.csv')
We will get an error:
ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines='skip' in order to skip bad lines:
pd.read_csv('test.csv', on_bad_lines='skip')
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
pd.read_csv('test.csv', on_bad_lines='warn')
warning like:
b'Skipping line 4: expected 3 fields, saw 4n'
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
sep—lineterminator—engine
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
import pandas as pd
pd.read_csv('test.csv', sep='t', lineterminator='rn')
Note that delimiter is an alias for sep.
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
cpythonpyarrow
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
pd.read_csv('test.csv', engine='python')
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
pd.read_csv('test.csv', skiprows=1, engine='python')
or skipping the last lines:
pd.read_csv('test.csv', skipfooter=1, engine='python')
Note that we need to use python as the read_csv() engine for parameter — skipfooter.
In some cases header=None can help in order to solve the error:
pd.read_csv('test.csv', header=None)
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
Dates
2022-03-20
2022-03-21
2022-03-22
Raw data
col_1,col_2,col_3
11,12,13
21,22,23
31,32,33
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
df_temp = pd.read_csv('test.csv', sep='@@', engine='python', names=['col'])
ix_last_start = df_temp[df_temp['col'].str.contains('# Raw data')].index.max()
result is:
4
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
df_analytics = pd.read_csv(file, skiprows=ix_last_start)
Conclusion
In this post we saw how to investigate the error:
ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
While reading a CSV file, you may get the “Pandas Error Tokenizing Data“. This mostly occurs due to the incorrect data in the CSV file.
You can solve python pandas error tokenizing data error by ignoring the offending lines using error_bad_lines=False.
In this tutorial, you’ll learn the cause and how to solve the error tokenizing data error.
If you’re in Hurry
You can use the below code snippet to solve the tokenizing error. You can solve the error by ignoring the offending lines and suppressing errors.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', error_bad_lines=False, engine ='python')
dfIf You Want to Understand Details, Read on…
In this tutorial, you’ll learn the causes for the exception “Error Tokenizing Data” and how it can be solved.
Cause of the Problem
- CSV file has two header lines
- Different separator is used
r– is a new line character and it is present in column names which makes subsequent column names to be read as next line- Lines of the CSV files have inconsistent number of columns
In the case of invalid rows which has an inconsistent number of columns, you’ll see an error as Expected 1 field in line 12, saw m. This means it expected only 1 field in the CSV file but it saw 12 values after tokenizing it. Hence, it doesn’t know how the tokenized values need to be handled. You can solve the errors by using one of the options below.
Finding the Problematic Line (Optional)
If you want to identify the line which is creating the problem while reading, you can use the below code snippet.
It uses the CSV reader. hence it is similar to the read_csv() method.
Snippet
import csv
with open("sample.csv", 'rb') as file_obj:
reader = csv.reader(file_obj)
line_no = 1
try:
for row in reader:
line_no += 1
except Exception as e:
print (("Error in the line number %d: %s %s" % (line_no, str(type(e)), e.message)))Using Err_Bad_Lines Parameter
When there is insufficient data in any of the rows, the tokenizing error will occur.
You can skip such invalid rows by using the err_bad_line parameter within the read_csv() method.
This parameter controls what needs to be done when a bad line occurs in the file being read.
If it’s set to,
False– Errors will be suppressed for Invalid linesTrue– Errors will be thrown for invalid lines
Use the below snippet to read the CSV file and ignore the invalid lines. Only a warning will be shown with the line number when there is an invalid lie found.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', error_bad_lines=False)
dfIn this case, the offending lines will be skipped and only the valid lines will be read from CSV and a dataframe will be created.
Using Python Engine
There are two engines supported in reading a CSV file. C engine and Python Engine.
C Engine
- Faster
- Uses C language to parse the CSV file
- Supports
float_precision - Cannot automatically detect the separator
- Doesn’t support skipping footer
Python Engine
- Slower when compared to C engine but its feature complete
- Uses Python language to parse the CSV file
- Doesn’t support
float_precision. Not required with Python - Can automatically detect the separator
- Supports skipping footer
Using the python engine can solve the problems faced while parsing the files.
For example, When you try to parse large CSV files, you may face the Error tokenizing data. c error out of memory. Using the python engine can solve the memory issues while parsing such big CSV files using the read_csv() method.
Use the below snippet to use the Python engine for reading the CSV file.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', engine='python', error_bad_lines=False)
dfThis is how you can use the python engine to parse the CSV file.
Optionally, this could also solve the error Error tokenizing data. c error out of memory when parsing the big CSV files.
Using Proper Separator
CSV files can have different separators such as tab separator or any other special character such as ;. In this case, an error will be thrown when reading the CSV file, if the default C engine is used.
You can parse the file successfully by specifying the separator explicitly using the sep parameter.
As an alternative, you can also use the python engine which will automatically detect the separator and parse the file accordingly.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', sep='t')
dfThis is how you can specify the separator explicitly which can solve the tokenizing errors while reading the CSV files.
Using Line Terminator
CSV file can contain r carriage return for separating the lines instead of the line separator n.
In this case, you’ll face CParserError: Error tokenizing data. C error: Buffer overflow caught - possible malformed input file when the line contains the r instead on n.
You can solve this error by using the line terminator explicitly using the lineterminator parameter.
Snippet
df = pd.read_csv('sample.csv',
lineterminator='n')This is how you can use the line terminator to parse the files with the terminator r.
Using header=None
CSV files can have incomplete headers which can cause tokenizing errors while parsing the file.
You can use header=None to ignore the first line headers while reading the CSV files.
This will parse the CSV file without headers and create a data frame. You can also add headers to column names by adding columns attribute to the read_csv() method.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, error_bad_lines=False)
dfThis is how you can ignore the headers which are incomplete and cause problems while reading the file.
Using Skiprows
CSV files can have headers in more than one row. This can happen when data is grouped into different sections and each group is having a name and has columns in each section.
In this case, you can ignore such rows by using the skiprows parameter. You can pass the number of rows to be skipped and the data will be read after skipping those number of rows.
Use the below snippet to skip the first two rows while reading the CSV file.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, skiprows=2, error_bad_lines=False)
dfThis is how you can skip or ignore the erroneous headers while reading the CSV file.
Reading As Lines and Separating
In a CSV file, you may have a different number of columns in each row. This can occur when some of the columns in the row are considered optional. You may need to parse such files without any problems during tokenizing.
In this case, you can read the file as lines and separate it later using the delimiter and create a dataframe out of it. This is helpful when you have varying lengths of rows.
In the below example,
- the file is read as lines by specifying the separator as a new line using
sep='n'. Now the file will be tokenized on each new line, and a single column will be available in the dataframe. - You can split the lines using the separator or regex and create different columns out of it.
expand=Trueexpands the split string into multiple columns.
Use the below snippet to read the file as lines and separate it using the separator.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, sep='n')
df = df[0].str.split('s|s', expand=True)
dfThis is how you can read the file as lines and later separate it to avoid problems while parsing the lines with an inconsistent number of columns.
Conclusion
To summarize, you’ve learned the causes of the Python Pandas Error tokenizing data and the different methods to solve it in different scenarios.
Different Errors while tokenizing data are,
Error tokenizing data. C error: Buffer overflow caught - possible malformed input fileParserError: Expected n fields in line x, saw mError tokenizing data. c error out of memory
Also learned the different engines available in the read_csv() method to parse the CSV file and the advantages and disadvantages of it.
You’ve also learned when to use the different methods appropriately.
If you have any questions, comment below.
You May Also Like
- How to write pandas dataframe to CSV
- How To Read An Excel File In Pandas – With Examples
- How To Read Excel With Multiple Sheets In Pandas?
- How To Solve Truth Value Of A Series Is Ambiguous. Use A.Empty, A.Bool(), A.Item(), A.Any() Or A.All()?
- How to solve xlrd.biffh.XLRDError: Excel xlsx file; not supported Error?
Содержание
- How to Solve Error Tokenizing Data on read_csv in Pandas
- Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
- Step 2: Use correct separator to solve Pandas tokenizing error
- Step 3: Use different engine for read_csv()
- Step 4: Identify the headers to solve Error Tokenizing Data
- Step 5: Autodetect skiprows for read_csv()
- Conclusion
- How To Fix pandas.parser.CParserError: Error tokenizing data
- Understanding why the error is raised and how to deal with it when reading CSV files in pandas
- Introduction
- Reproducing the error
- Fixing the file manually
- Specifying line terminator
- Specifying the correct delimiter and headers
- Skipping rows
- Final Thoughts
- Python ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z (Example)
- Example Data & Software Libraries
- Reproduce the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
- Debug the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
- Video & Further Resources
- How to Solve Python Pandas Error Tokenizing Data Error?
- Cause of the Problem
- Finding the Problematic Line (Optional)
- Using Err_Bad_Lines Parameter
- Using Python Engine
- Using Proper Separator
- Using Line Terminator
- Using header=None
- Using Skiprows
- Reading As Lines and Separating
- Conclusion
How to Solve Error Tokenizing Data on read_csv in Pandas
In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
which we are going to read by — read_csv() method:
We will get an error:
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines=’skip’ in order to skip bad lines:
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
Note that delimiter is an alias for sep .
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
or skipping the last lines:
Note that we need to use python as the read_csv() engine for parameter — skipfooter .
In some cases header=None can help in order to solve the error:
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
Conclusion
In this post we saw how to investigate the error:
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
Источник
How To Fix pandas.parser.CParserError: Error tokenizing data
Understanding why the error is raised and how to deal with it when reading CSV files in pandas
Introduction
Importing data from csv files is probably the most commonly used way of instantiating pandas DataFrames. However, many times this could be a little bit tricky especially when the data included in the file are not in the expected form. In these cases, pandas parser may raise an error similar to the one reported below:
In today’s short guide we will discuss why this error is being raised in the first place and additionally, we will discuss a few ways that could eventually help you deal with it.
Reproducing the error
First, let’s try to reproduce the error using a small dataset I have prepared as part of this tutorial.
Now if we attempt to read in the file using read_csv :
we are going to get the following error
The error is pretty clear as it indicates that on the 4th line instead of 4, 6 fields were observed (and by the way, the same issue occurs in the last line as well).
By default, read_csv uses comma ( , ) as the delimiter but clearly, two lines in the file have more separators than expected. The expected number in this occasion is actually 4, since our headers (i.e. the first line of the file) contains 4 fields separated by commas.
Fixing the file manually
The most obvious solution to the problem, is to fix the data file manually by removing the extra separators in the lines causing us troubles. This is actually the best solution (assuming that you have specified the right delimiters, headers etc. when calling read_csv function). However, this may be quite tricky and painful when you need to deal with large files containing thousands of lines.
Specifying line terminator
Another cause of this error may be related to some carriage returns (i.e. ‘r’ ) in the data. In some occasions this is actually introduced by pandas.to_csv() method. When writing pandas DataFrames to CSV files, a carriage return is added to column names in which the method will then write the subsequent column names to the first column of the pandas DataFrame. And thus we’ll end up with different number of columns in the first rows.
If that’s the case, then you can explicitly specify the line terminator to ‘n’ using the corresponding parameter when calling read_csv() :
Specifying the correct delimiter and headers
The error may also be related to the delimiters and/or headers (not) specified when calling read_csv . Make sure to pass both the correct separator and headers.
For example, the arguments below specify that ; is the delimiter used to separate columns (by default commas are used as delimiters) and that the file does not contain any headers at all.
Skipping rows
Skipping rows that are causing the error should be your last resort and I would personally discourage you from doing so, but I guess there are certain use cases where this may be acceptable.
If that’s the case, then you can do so by setting error_bad_lines to False when calling read_csv function:
As you can see from the output above, the lines that they were causing errors were actually skipped and we can now move on with whatever we’d like to do with our pandas DataFrame.
Final Thoughts
In today’s short guide, we discussed a few cases where pandas.errors.ParserError: Error tokenizing data is raised by the pandas parser when reading csv files into pandas DataFrames.
Additionally, we showcased how to deal with the error by fixing the errors or typos in the data file itself, or by specifying the appropriate line terminator. Finally, we also discussed how to skip lines causing errors but keep in mind that in most of the cases this should be avoided.
Become a member and read every story on Medium. Your membership fee directly supports me and other writers you read. You’ll also get full access to every story on Medium.
Источник
Python ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z (Example)
In this tutorial you’ll learn how to fix the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language.
The article consists of the following information:
Let’s get started.
Example Data & Software Libraries
Consider the CSV file illustrated below as a basis for this tutorial:
You may already note that rows 4 and 6 contain one value too much. Those two rows contain four different values, but the other rows contain only three values.
Let’s assume that we want to read this CSV file as a pandas DataFrame into Python.
For this, we first have to import the pandas library:
import pandas as pd # Load pandas
Let’s move on to the examples!
Reproduce the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
In this section, I’ll show how to replicate the error message “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z”.
Let’s assume that we want to read our example CSV file using the default settings of the read_csv function. Then, we might try to import our data as shown below:
data_import = pd.read_csv(‘data.csv’) # Try to import CSV file # ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
Unfortunately, the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” is returned after executing the Python syntax above.
The reason for this is that our CSV file contains too many values in some of the rows.
In the next section, I’ll show an easy solution for this problem. So keep on reading…
Debug the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
In this example, I’ll explain an easy fix for the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language.
We can ignore all lines in our CSV file that are formatted wrongly by specifying the error_bad_lines argument to False.
Have a look at the example code below:
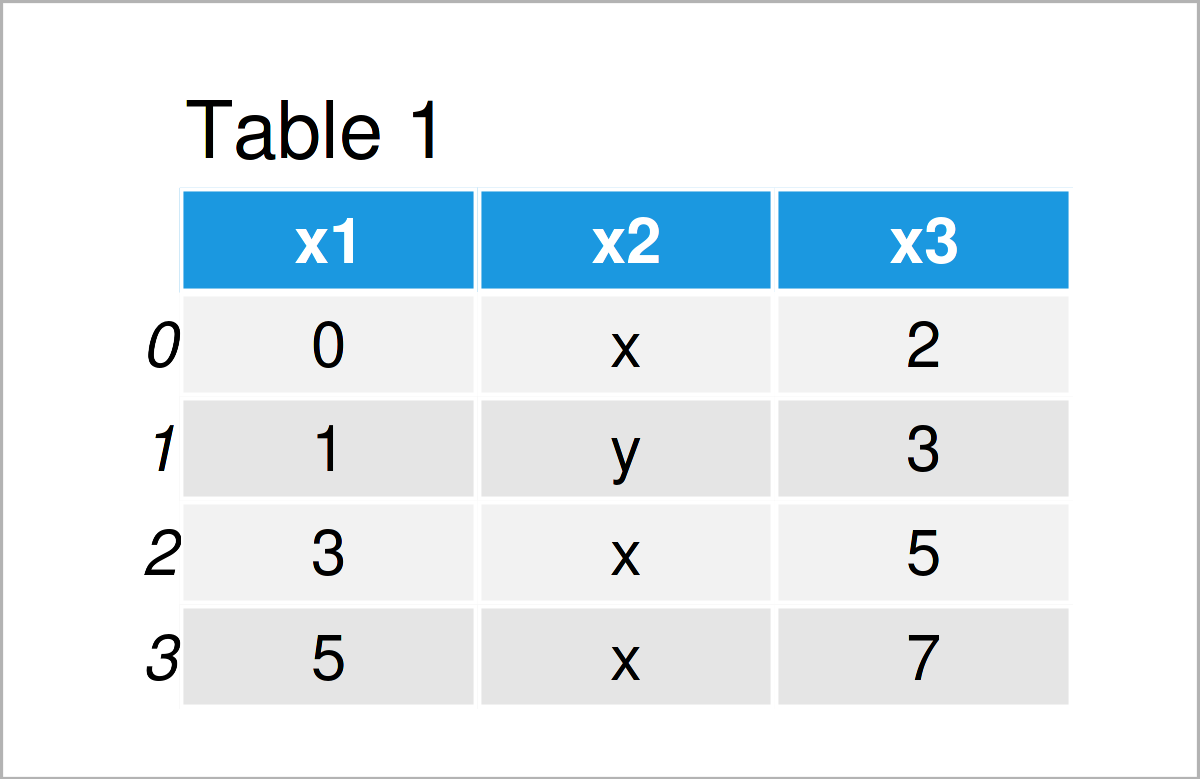
data_import = pd.read_csv(‘data.csv’, # Remove rows with errors error_bad_lines = False) print(data_import) # Print imported pandas DataFrame
As shown in Table 2, we have created a valid pandas DataFrame output using the previous code. As you can see, we have simply skipped the rows with too many values.
This is a simply trick that usually works. However, please note that this trick should be done with care, since the discussed error message typically points to more general issues with your data.
For that reason, it’s advisable to investigate why some of the rows are not formatted properly.
For this, I can also recommend this thread on Stack Overflow. It discusses how to identify wrong lines, and it also discusses other less common reasons for the error message “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z”.
Video & Further Resources
Have a look at the following video on my YouTube channel. In the video, I’m explaining the Python codes of this tutorial:
The YouTube video will be added soon.
Furthermore, you might read the other articles on this website. You can find some interesting tutorials below:
Summary: In this article, I have explained how to handle the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language. If you have any further questions or comments, let me know in the comments. Furthermore, don’t forget to subscribe to my email newsletter to get updates on new articles.
Источник
How to Solve Python Pandas Error Tokenizing Data Error?
While reading a CSV file, you may get the “Pandas Error Tokenizing Data“. This mostly occurs due to the incorrect data in the CSV file.
You can solve python pandas error tokenizing data error by ignoring the offending lines using error_bad_lines=False .
In this tutorial, you’ll learn the cause and how to solve the error tokenizing data error.
If you’re in Hurry
You can use the below code snippet to solve the tokenizing error. You can solve the error by ignoring the offending lines and suppressing errors.
Snippet
If You Want to Understand Details, Read on…
In this tutorial, you’ll learn the causes for the exception “Error Tokenizing Data” and how it can be solved.
Table of Contents
Cause of the Problem
- CSV file has two header lines
- Different separator is used
- r – is a new line character and it is present in column names which makes subsequent column names to be read as next line
- Lines of the CSV files have inconsistent number of columns
In the case of invalid rows which has an inconsistent number of columns, you’ll see an error as Expected 1 field in line 12, saw m . This means it expected only 1 field in the CSV file but it saw 12 values after tokenizing it. Hence, it doesn’t know how the tokenized values need to be handled. You can solve the errors by using one of the options below.
Finding the Problematic Line (Optional)
If you want to identify the line which is creating the problem while reading, you can use the below code snippet.
It uses the CSV reader. hence it is similar to the read_csv() method.
Snippet
Using Err_Bad_Lines Parameter
When there is insufficient data in any of the rows, the tokenizing error will occur.
You can skip such invalid rows by using the err_bad_line parameter within the read_csv() method.
This parameter controls what needs to be done when a bad line occurs in the file being read.
- False – Errors will be suppressed for Invalid lines
- True – Errors will be thrown for invalid lines
Use the below snippet to read the CSV file and ignore the invalid lines. Only a warning will be shown with the line number when there is an invalid lie found.
Snippet
In this case, the offending lines will be skipped and only the valid lines will be read from CSV and a dataframe will be created.
Using Python Engine
There are two engines supported in reading a CSV file. C engine and Python Engine.
C Engine
- Faster
- Uses C language to parse the CSV file
- Supports float_precision
- Cannot automatically detect the separator
- Doesn’t support skipping footer
Python Engine
- Slower when compared to C engine but its feature complete
- Uses Python language to parse the CSV file
- Doesn’t support float_precision . Not required with Python
- Can automatically detect the separator
- Supports skipping footer
Using the python engine can solve the problems faced while parsing the files.
For example, When you try to parse large CSV files, you may face the Error tokenizing data. c error out of memory. Using the python engine can solve the memory issues while parsing such big CSV files using the read_csv() method.
Use the below snippet to use the Python engine for reading the CSV file.
Snippet
This is how you can use the python engine to parse the CSV file.
Optionally, this could also solve the error Error tokenizing data. c error out of memory when parsing the big CSV files.
Using Proper Separator
CSV files can have different separators such as tab separator or any other special character such as ; . In this case, an error will be thrown when reading the CSV file, if the default C engine is used.
You can parse the file successfully by specifying the separator explicitly using the sep parameter.
As an alternative, you can also use the python engine which will automatically detect the separator and parse the file accordingly.
Snippet
This is how you can specify the separator explicitly which can solve the tokenizing errors while reading the CSV files.
Using Line Terminator
CSV file can contain r carriage return for separating the lines instead of the line separator n .
In this case, you’ll face CParserError: Error tokenizing data. C error: Buffer overflow caught — possible malformed input file when the line contains the r instead on n .
You can solve this error by using the line terminator explicitly using the lineterminator parameter.
Snippet
This is how you can use the line terminator to parse the files with the terminator r .
CSV files can have incomplete headers which can cause tokenizing errors while parsing the file.
You can use header=None to ignore the first line headers while reading the CSV files.
This will parse the CSV file without headers and create a data frame. You can also add headers to column names by adding columns attribute to the read_csv() method.
Snippet
This is how you can ignore the headers which are incomplete and cause problems while reading the file.
Using Skiprows
CSV files can have headers in more than one row. This can happen when data is grouped into different sections and each group is having a name and has columns in each section.
In this case, you can ignore such rows by using the skiprows parameter. You can pass the number of rows to be skipped and the data will be read after skipping those number of rows.
Use the below snippet to skip the first two rows while reading the CSV file.
Snippet
This is how you can skip or ignore the erroneous headers while reading the CSV file.
Reading As Lines and Separating
In a CSV file, you may have a different number of columns in each row. This can occur when some of the columns in the row are considered optional. You may need to parse such files without any problems during tokenizing.
In this case, you can read the file as lines and separate it later using the delimiter and create a dataframe out of it. This is helpful when you have varying lengths of rows.
In the below example,
- the file is read as lines by specifying the separator as a new line using sep=’n’ . Now the file will be tokenized on each new line, and a single column will be available in the dataframe.
- You can split the lines using the separator or regex and create different columns out of it.
- expand=True expands the split string into multiple columns.
Use the below snippet to read the file as lines and separate it using the separator.
Snippet
This is how you can read the file as lines and later separate it to avoid problems while parsing the lines with an inconsistent number of columns.
Conclusion
To summarize, you’ve learned the causes of the Python Pandas Error tokenizing data and the different methods to solve it in different scenarios.
Different Errors while tokenizing data are,
- Error tokenizing data. C error: Buffer overflow caught — possible malformed input file
- ParserError: Expected n fields in line x, saw m
- Error tokenizing data. c error out of memory
Also learned the different engines available in the read_csv() method to parse the CSV file and the advantages and disadvantages of it.
You’ve also learned when to use the different methods appropriately.
Источник
Recently, I burned about 3 hours trying to load a large CSV file into Python Pandas using the read_csv function, only to consistently run into the following error:
ParserError Traceback (most recent call last) <ipython-input-6-b51ad8562823> in <module>() ... pandas_libsparsers.pyx in pandas._libs.parsers.TextReader.read (pandas_libsparsers.c:10862)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_low_memory (pandas_libsparsers.c:11138)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_rows (pandas_libsparsers.c:11884)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._tokenize_rows (pandas_libsparsers.c:11755)() pandas_libsparsers.pyx in pandas._libs.parsers.raise_parser_error (pandas_libsparsers.c:28765)() Error tokenizing data. C error: EOF inside string starting at line XXXX
“Error tokenising data. C error: EOF inside string starting at line”.
There was an erroneous character about 5000 lines into the CSV file that prevented the Pandas CSV parser from reading the entire file. Excel had no problems opening the file, and no amount of saving/re-saving/changing encodings was working. Manually removing the offending line worked, but ultimately, another character 6000 lines further into the file caused the same issue.
The solution was to use the parameter engine=’python’ in the read_csv function call. The Pandas CSV parser can use two different “engines” to parse CSV file – Python or C (default).
pandas.read_csv(filepath_or_buffer, sep=', ', delimiter=None,
header='infer', names=None,
index_col=None, usecols=None, squeeze=False,
..., engine=None, ...)
engine : {‘c’, ‘python’}, optional
Parser engine to use. The C engine is faster while the python engine is currently more feature-complete.
The Python engine is described as “slower, but more feature complete” in the Pandas documentation. However, in this case, the python engine sorts the problem, without a massive time impact (overall file size was approx 200,000 rows).
UnicodeDecodeError
The other big problem with reading in CSV files that I come across is the error:
“UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
Character encoding issues can be usually fixed by opening the CSV file in Sublime Text, and then “Save with encoding” choosing UTF-8. Then adding the encoding=’utf8′ to the pandas.read_csv command allows Pandas to open the CSV file without trouble. This error appears regularly for me with files saved in Excel.
encoding : str, default None
@WillAyd Let me know if you need additional info.
Since GitHub doesn’t accept CSVs, I changed the extension to .txt.
Here’s the code which will trigger the exception.
for chunk in pandas.read_csv('debug.csv', chunksize=1000, names=range(2504)): pass
Here’s the file: debug.txt
Here’s the exception from Windows 10, using Anaconda.
Python 3.6.5 |Anaconda, Inc.| (default, Mar 29 2018, 13:32:41) [MSC v.1900 64 bit (AMD64)] on win32
Type "help", "copyright", "credits" or "license" for more information.
>>> import pandas
>>> for chunk in pandas.read_csv('debug.csv', chunksize=1000, names=range(2504)): pass
...
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "D:programsanaconda3libsite-packagespandasioparsers.py", line 1007, in __next__
return self.get_chunk()
File "D:programsanaconda3libsite-packagespandasioparsers.py", line 1070, in get_chunk
return self.read(nrows=size)
File "D:programsanaconda3libsite-packagespandasioparsers.py", line 1036, in read
ret = self._engine.read(nrows)
File "D:programsanaconda3libsite-packagespandasioparsers.py", line 1848, in read
data = self._reader.read(nrows)
File "pandas_libsparsers.pyx", line 876, in pandas._libs.parsers.TextReader.read
File "pandas_libsparsers.pyx", line 903, in pandas._libs.parsers.TextReader._read_low_memory
File "pandas_libsparsers.pyx", line 945, in pandas._libs.parsers.TextReader._read_rows
File "pandas_libsparsers.pyx", line 932, in pandas._libs.parsers.TextReader._tokenize_rows
File "pandas_libsparsers.pyx", line 2112, in pandas._libs.parsers.raise_parser_error
pandas.errors.ParserError: Error tokenizing data. C error: Buffer overflow caught - possible malformed input file.
And the same on RedHat.
$ python3
Python 3.6.6 (default, Aug 13 2018, 18:24:23)
[GCC 4.8.5 20150623 (Red Hat 4.8.5-28)] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>> import pandas
>>> for chunk in pandas.read_csv('debug.csv', chunksize=1000, names=range(2504)): pass
...
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/usr/lib64/python3.6/site-packages/pandas/io/parsers.py", line 1007, in __next__
return self.get_chunk()
File "/usr/lib64/python3.6/site-packages/pandas/io/parsers.py", line 1070, in get_chunk
return self.read(nrows=size)
File "/usr/lib64/python3.6/site-packages/pandas/io/parsers.py", line 1036, in read
ret = self._engine.read(nrows)
File "/usr/lib64/python3.6/site-packages/pandas/io/parsers.py", line 1848, in read
data = self._reader.read(nrows)
File "pandas/_libs/parsers.pyx", line 876, in pandas._libs.parsers.TextReader.read
File "pandas/_libs/parsers.pyx", line 903, in pandas._libs.parsers.TextReader._read_low_memory
File "pandas/_libs/parsers.pyx", line 945, in pandas._libs.parsers.TextReader._read_rows
File "pandas/_libs/parsers.pyx", line 932, in pandas._libs.parsers.TextReader._tokenize_rows
File "pandas/_libs/parsers.pyx", line 2112, in pandas._libs.parsers.raise_parser_error
pandas.errors.ParserError: Error tokenizing data. C error: Buffer overflow caught - possible malformed input file.
Common reasons for ParserError: Error tokenizing data when initiating a Pandas DataFrame include:
-
Using the wrong delimiter
-
Number of fields in certain rows do not match with header
To resolve the error we can try the following:
-
Specifying the delimiter through
sepparameter inread_csv(~) -
Fixing the original source file
-
Skipping bad rows
Examples
Specifying sep
By default the read_csv(~) method assumes sep=",". Therefore when reading files that use a different delimiter, make sure to explicitly specify the delimiter to use.
Consider the following slash-delimited file called test.txt:
col1/col2/col3
1/A/4
2/B/5
3/C,D,E/6
To initialize a DataFrame using default sep=",":
import pandas as pd
df = pd.read_csv('test.txt')
ParserError: Error tokenizing data. C error: Expected 1 fields in line 4, saw 3
An error is raised as the first line in the file does not contain any commas, so read_csv(~) expects all lines in the file to only contain 1 field. However, line 4 has 3 fields as it contains two commas, which results in the ParserError.
To initialize the DataFrame by correctly specifying slash (/) as the delimiter:
df = pd.read_csv('test.txt', sep='/')
df
col1 col2 col3
0 1 A 4
1 2 B 5
2 3 C,D,E 6
We can now see the DataFrame is initialized as expected with each line containing the 3 fields which were separated by slashes (/) in the original file.
Fixing original source file
Consider the following comma-separated file called test.csv:
col1,col2,col3
1,A,4
2,B,5,
3,C,6
To initialize a DataFrame using the above file:
import pandas as pd
df = pd.read_csv('test.csv')
ParserError: Error tokenizing data. C error: Expected 3 fields in line 3, saw 4
Here there is an error on the 3rd line as 4 fields are observed instead of 3, caused by the additional comma at the end of the line.
To resolve this error, we can correct the original file by removing the extra comma at the end of line 3:
col1,col2,col3
1,A,4
2,B,5
3,C,6
To now initialize the DataFrame again using the corrected file:
df = pd.read_csv('test.csv')
df
col1 col2 col3
0 1 A 4
1 2 B 5
2 3 C 6
Skipping bad rows
Consider the following comma-separated file called test.csv:
col1,col2,col3
1,A,4
2,B,5,
3,C,6
To initialize a DataFrame using the above file:
import pandas as pd
df = pd.read_csv('test.csv')
ParserError: Error tokenizing data. C error: Expected 3 fields in line 3, saw 4
Here there is an error on the 3rd line as 4 fields are observed instead of 3, caused by the additional comma at the end of the line.
To skip bad rows pass on_bad_lines='skip' to read_csv(~):
df = pd.read_csv('test.csv', on_bad_lines='skip')
df
col1 col2 col3
0 1 A 4
1 3 C 6
Notice how the problematic third line in the original file has been skipped in the resulting DataFrame.
WARNING
This should be your last resort as valuable information could be contained within the problematic lines. Skipping these rows means you lose this information. As much as possible try to identify the root cause of the error and fix the underlying problem.