Computes the mean of absolute difference between labels and predictions.
Inherits From: Loss
View aliases
Main aliases
tf.losses.MeanAbsoluteError
Compat aliases for migration
See
Migration guide for
more details.
tf.compat.v1.keras.losses.MeanAbsoluteError
tf.keras.losses.MeanAbsoluteError(
reduction=losses_utils.ReductionV2.AUTO,
name='mean_absolute_error'
)
Used in the notebooks
| Used in the tutorials |
|---|
|
loss = abs(y_true - y_pred)
Standalone usage:
y_true = [[0., 1.], [0., 0.]]y_pred = [[1., 1.], [1., 0.]]# Using 'auto'/'sum_over_batch_size' reduction type.mae = tf.keras.losses.MeanAbsoluteError()mae(y_true, y_pred).numpy()0.5
# Calling with 'sample_weight'.mae(y_true, y_pred, sample_weight=[0.7, 0.3]).numpy()0.25
# Using 'sum' reduction type.mae = tf.keras.losses.MeanAbsoluteError(reduction=tf.keras.losses.Reduction.SUM)mae(y_true, y_pred).numpy()1.0
# Using 'none' reduction type.mae = tf.keras.losses.MeanAbsoluteError(reduction=tf.keras.losses.Reduction.NONE)mae(y_true, y_pred).numpy()array([0.5, 0.5], dtype=float32)
Usage with the compile() API:
model.compile(optimizer='sgd', loss=tf.keras.losses.MeanAbsoluteError())
Args |
|
|---|---|
reduction
|
Type of tf.keras.losses.Reduction to apply toloss. Default value is AUTO. AUTO indicates that the reductionoption will be determined by the usage context. For almost all cases this defaults to SUM_OVER_BATCH_SIZE. When used withtf.distribute.Strategy, outside of built-in training loops such astf.keras compile and fit, using AUTO orSUM_OVER_BATCH_SIZE will raise an error. Please see this customtraining tutorial for more details. |
name
|
Optional name for the instance. Defaults to ‘mean_absolute_error’. |
Methods
from_config
View source
@classmethodfrom_config( config )
Instantiates a Loss from its config (output of get_config()).
| Args | |
|---|---|
config
|
Output of get_config().
|
| Returns |
|---|
A keras.losses.Loss instance.
|
get_config
View source
get_config()
Returns the config dictionary for a Loss instance.
__call__
View source
__call__(
y_true, y_pred, sample_weight=None
)
Invokes the Loss instance.
| Args | |
|---|---|
y_true
|
Ground truth values. shape = [batch_size, d0, .. dN], exceptsparse loss functions such as sparse categorical crossentropy where shape = [batch_size, d0, .. dN-1]
|
y_pred
|
The predicted values. shape = [batch_size, d0, .. dN]
|
sample_weight
|
Optional sample_weight acts as a coefficient for theloss. If a scalar is provided, then the loss is simply scaled by the given value. If sample_weight is a tensor of size [batch_size],then the total loss for each sample of the batch is rescaled by the corresponding element in the sample_weight vector. If the shape ofsample_weight is [batch_size, d0, .. dN-1] (or can bebroadcasted to this shape), then each loss element of y_pred isscaled by the corresponding value of sample_weight. (Noteon dN-1: all loss functions reduce by 1 dimension, usuallyaxis=-1.) |
| Returns |
|---|
Weighted loss float Tensor. If reduction is NONE, this hasshape [batch_size, d0, .. dN-1]; otherwise, it is scalar. (NotedN-1 because all loss functions reduce by 1 dimension, usuallyaxis=-1.) |
| Raises | |
|---|---|
ValueError
|
If the shape of sample_weight is invalid.
|
Использование функций потерь
Функция потерь (или объективная функция, или функция оценки результатов оптимизации) является одним из двух параметров, необходимых для компиляции модели:
model.compile(loss=’mean_squared_error’, optimizer=’sgd’)
from keras import losses
model.compile(loss=losses.mean_squared_error, optimizer=’sgd’)
Можно либо передать имя существующей функции потерь, либо передать символическую функцию TensorFlow/Theano, которая возвращает скаляр для каждой точки данных и принимает следующие два аргумента:
y_true: истинные метки. Тензор TensorFlow/Theano.
y_pred: Прогнозы. Тензор TensorFlow/Theano той же формы, что и y_true.
Фактически оптимизированная цель — это среднее значение выходного массива по всем точкам данных.
Доступные функции потери
mean_squared_error
keras.losses.mean_squared_error(y_true, y_pred)
mean_absolute_error
keras.losses.mean_absolute_error(y_true, y_pred)
mean_absolute_percentage_error
keras.losses.mean_absolute_percentage_error(y_true, y_pred)
mean_squared_logarithmic_error
keras.losses.mean_squared_logarithmic_error(y_true, y_pred)
squared_hinge
keras.losses.squared_hinge(y_true, y_pred)
hinge
keras.losses.hinge(y_true, y_pred)
categorical_hinge
keras.losses.categorical_hinge(y_true, y_pred)
logcosh
keras.losses.logcosh(y_true, y_pred)
Логарифм гиперболического косинуса ошибки прогнозирования.
log(cosh(x)) приблизительно равен (x ** 2) / 2 для малого x и abs(x) — log(2) для большого x. Это означает, что ‘logcosh’ работает в основном как средняя квадратичная ошибка, но не будет так сильно зависеть от случайного сильно неправильного предсказания.
Аргументы
- y_true: тензор истинных целей.
- y_pred: тензор прогнозируемых целей.
Возвращает
Тензор с одной записью о скалярной потере на каждый сэмпл.
huber_loss
keras.losses.huber_loss(y_true, y_pred, delta=1.0)
categorical_crossentropy
keras.losses.categorical_crossentropy(y_true, y_pred, from_logits=False, label_smoothing=0)
sparse_categorical_crossentropy
keras.losses.sparse_categorical_crossentropy(y_true, y_pred, from_logits=False, axis=-1)
binary_crossentropy
keras.losses.binary_crossentropy(y_true, y_pred, from_logits=False, label_smoothing=0)
kullback_leibler_divergence
keras.losses.kullback_leibler_divergence(y_true, y_pred)
poisson
keras.losses.poisson(y_true, y_pred)
cosine_proximity
keras.losses.cosine_proximity(y_true, y_pred, axis=-1)
is_categorical_crossentropy
keras.losses.is_categorical_crossentropy(loss)
Примечание: при использовании потери categorical_crossentropy ваши данные должны быть в категориальном формате (например, если у вас 10 классов, то целью для каждой выборки должен быть 10-мерный вектор, который является полностью нулевым, за исключением 1 в индексе, соответствующем классу выборки). Для того, чтобы преобразовать целые данные в категорические, можно использовать утилиту Keras to_categorical:
from keras.utils import to_categorical
categorical_labels = to_categorical(int_labels, num_classes=None)
При использовании переменной sparse_categorical_crossentropy loss, ваши данные должны быть целыми. Если у вас есть категориальные данные, следует использовать categoryical_crossentropy.
categoryical_crossentropy — это еще один термин для обозначения потери лога по нескольким классам.
There are many definitions for a regression problem but in our case, we’re going to simplify it to be: predicting a number.
For example, you might want to:
- Predict the selling price of houses given information about them (such as number of rooms, size, number of bathrooms).
- Predict the coordinates of a bounding box of an item in an image.
- Predict the cost of medical insurance for an individual given their demographics (age, sex, gender, race).
In this notebook, we’re going to set the foundations for how you can take a sample of inputs (this is your data), build a neural network to discover patterns in those inputs and then make a prediction (in the form of a number) based on those inputs.
What we’re going to cover¶
Specifically, we’re going to go through doing the following with TensorFlow:
- Architecture of a regression model
- Input shapes and output shapes
X: features/data (inputs)y: labels (outputs)
- Creating custom data to view and fit
- Steps in modelling
- Creating a model
- Compiling a model
- Defining a loss function
- Setting up an optimizer
- Creating evaluation metrics
- Fitting a model (getting it to find patterns in our data)
- Evaluating a model
- Visualizng the model («visualize, visualize, visualize»)
- Looking at training curves
- Compare predictions to ground truth (using our evaluation metrics)
- Saving a model (so we can use it later)
- Loading a model
Don’t worry if none of these make sense now, we’re going to go through each.
How you can use this notebook¶
You can read through the descriptions and the code (it should all run), but there’s a better option.
Write all of the code yourself.
Yes. I’m serious. Create a new notebook, and rewrite each line by yourself. Investigate it, see if you can break it, why does it break?
You don’t have to write the text descriptions but writing the code yourself is a great way to get hands-on experience.
Don’t worry if you make mistakes, we all do. The way to get better and make less mistakes is to write more code.
Typical architecture of a regresison neural network¶
The word typical is on purpose.
Why?
Because there are many different ways (actually, there’s almost an infinite number of ways) to write neural networks.
But the following is a generic setup for ingesting a collection of numbers, finding patterns in them and then outputing some kind of target number.
Yes, the previous sentence is vague but we’ll see this in action shortly.
| Hyperparameter | Typical value |
|---|---|
| Input layer shape | Same shape as number of features (e.g. 3 for # bedrooms, # bathrooms, # car spaces in housing price prediction) |
| Hidden layer(s) | Problem specific, minimum = 1, maximum = unlimited |
| Neurons per hidden layer | Problem specific, generally 10 to 100 |
| Output layer shape | Same shape as desired prediction shape (e.g. 1 for house price) |
| Hidden activation | Usually ReLU (rectified linear unit) |
| Output activation | None, ReLU, logistic/tanh |
| Loss function | MSE (mean square error) or MAE (mean absolute error)/Huber (combination of MAE/MSE) if outliers |
| Optimizer | SGD (stochastic gradient descent), Adam |
Table 1: Typical architecture of a regression network. Source: Adapted from page 293 of Hands-On Machine Learning with Scikit-Learn, Keras & TensorFlow Book by Aurélien Géron
Again, if you’re new to neural networks and deep learning in general, much of the above table won’t make sense. But don’t worry, we’ll be getting hands-on with all of it soon.
🔑 Note: A hyperparameter in machine learning is something a data analyst or developer can set themselves, where as a parameter usually describes something a model learns on its own (a value not explicitly set by an analyst).
Okay, enough talk, let’s get started writing code.
To use TensorFlow, we’ll import it as the common alias tf (short for TensorFlow).
In [18]:
import tensorflow as tf print(tf.__version__) # check the version (should be 2.x+)
import tensorflow as tf
print(tf.__version__) # check the version (should be 2.x+)
Creating data to view and fit¶
Since we’re working on a regression problem (predicting a number) let’s create some linear data (a straight line) to model.
In [19]:
import numpy as np import matplotlib.pyplot as plt # Create features X = np.array([-7.0, -4.0, -1.0, 2.0, 5.0, 8.0, 11.0, 14.0]) # Create labels y = np.array([3.0, 6.0, 9.0, 12.0, 15.0, 18.0, 21.0, 24.0]) # Visualize it plt.scatter(X, y);
import numpy as np
import matplotlib.pyplot as plt
# Create features
X = np.array([-7.0, -4.0, -1.0, 2.0, 5.0, 8.0, 11.0, 14.0])
# Create labels
y = np.array([3.0, 6.0, 9.0, 12.0, 15.0, 18.0, 21.0, 24.0])
# Visualize it
plt.scatter(X, y);
Before we do any modelling, can you calculate the pattern between X and y?
For example, say I asked you, based on this data what the y value would be if X was 17.0?
Or how about if X was -10.0?
This kind of pattern discovery is the essence of what we’ll be building neural networks to do for us.
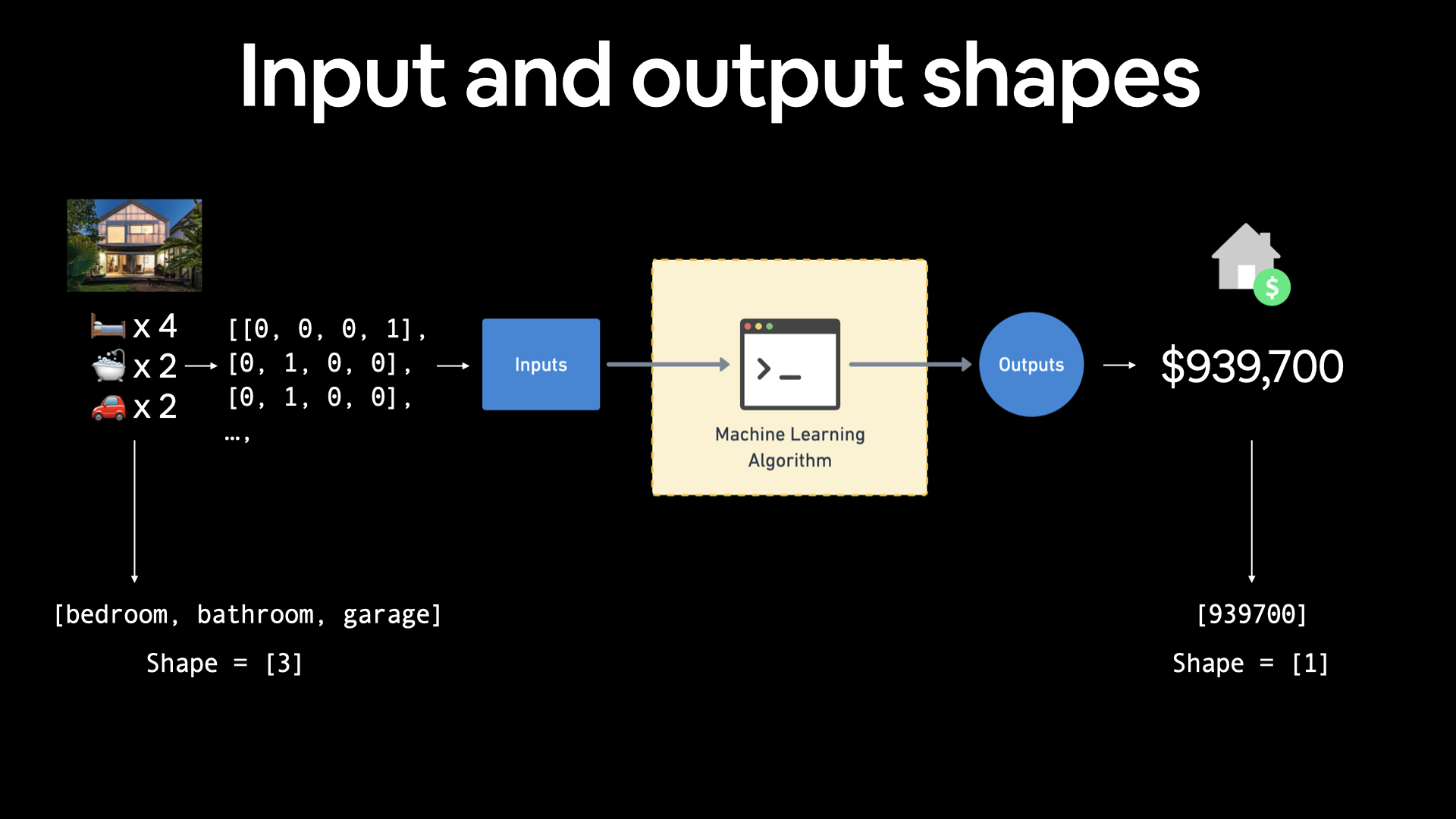
Regression input shapes and output shapes¶
One of the most important concepts when working with neural networks are the input and output shapes.
The input shape is the shape of your data that goes into the model.
The output shape is the shape of your data you want to come out of your model.
These will differ depending on the problem you’re working on.
Neural networks accept numbers and output numbers. These numbers are typically represented as tensors (or arrays).
Before, we created data using NumPy arrays, but we could do the same with tensors.
In [20]:
# Example input and output shapes of a regresson model house_info = tf.constant(["bedroom", "bathroom", "garage"]) house_price = tf.constant([939700]) house_info, house_price
# Example input and output shapes of a regresson model
house_info = tf.constant([«bedroom», «bathroom», «garage»])
house_price = tf.constant([939700])
house_info, house_price
Out[20]:
(<tf.Tensor: shape=(3,), dtype=string, numpy=array([b'bedroom', b'bathroom', b'garage'], dtype=object)>, <tf.Tensor: shape=(1,), dtype=int32, numpy=array([939700], dtype=int32)>)
In [22]:
import numpy as np import matplotlib.pyplot as plt # Create features (using tensors) X = tf.constant([-7.0, -4.0, -1.0, 2.0, 5.0, 8.0, 11.0, 14.0]) # Create labels (using tensors) y = tf.constant([3.0, 6.0, 9.0, 12.0, 15.0, 18.0, 21.0, 24.0]) # Visualize it plt.scatter(X, y);
import numpy as np
import matplotlib.pyplot as plt
# Create features (using tensors)
X = tf.constant([-7.0, -4.0, -1.0, 2.0, 5.0, 8.0, 11.0, 14.0])
# Create labels (using tensors)
y = tf.constant([3.0, 6.0, 9.0, 12.0, 15.0, 18.0, 21.0, 24.0])
# Visualize it
plt.scatter(X, y);
Our goal here will be to use X to predict y.
So our input will be X and our output will be y.
Knowing this, what do you think our input and output shapes will be?
Let’s take a look.
In [23]:
# Take a single example of X input_shape = X[0].shape # Take a single example of y output_shape = y[0].shape input_shape, output_shape # these are both scalars (no shape)
# Take a single example of X
input_shape = X[0].shape
# Take a single example of y
output_shape = y[0].shape
input_shape, output_shape # these are both scalars (no shape)
Out[23]:
(TensorShape([]), TensorShape([]))
Huh?
From this it seems our inputs and outputs have no shape?
How could that be?
It’s because no matter what kind of data we pass to our model, it’s always going to take as input and return as ouput some kind of tensor.
But in our case because of our dataset (only 2 small lists of numbers), we’re looking at a special kind of tensor, more specifically a rank 0 tensor or a scalar.
In [24]:
# Let's take a look at the single examples invidually X[0], y[0]
# Let’s take a look at the single examples invidually
X[0], y[0]
Out[24]:
(<tf.Tensor: shape=(), dtype=float32, numpy=-7.0>, <tf.Tensor: shape=(), dtype=float32, numpy=3.0>)
In our case, we’re trying to build a model to predict the pattern between X[0] equalling -7.0 and y[0] equalling 3.0.
So now we get our answer, we’re trying to use 1 X value to predict 1 y value.
You might be thinking, «this seems pretty complicated for just predicting a straight line…».
And you’d be right.
But the concepts we’re covering here, the concepts of input and output shapes to a model are fundamental.
In fact, they’re probably two of the things you’ll spend the most time on when you work with neural networks: making sure your input and outputs are in the correct shape.
If it doesn’t make sense now, we’ll see plenty more examples later on (soon you’ll notice the input and output shapes can be almost anything you can imagine).
If you were working on building a machine learning algorithm for predicting housing prices, your inputs may be number of bedrooms, number of bathrooms and number of garages, giving you an input shape of 3 (3 different features). And since you’re trying to predict the price of the house, your output shape would be 1.
Steps in modelling with TensorFlow¶
Now we know what data we have as well as the input and output shapes, let’s see how we’d build a neural network to model it.
In TensorFlow, there are typically 3 fundamental steps to creating and training a model.
- Creating a model — piece together the layers of a neural network yourself (using the Functional or Sequential API) or import a previously built model (known as transfer learning).
- Compiling a model — defining how a models performance should be measured (loss/metrics) as well as defining how it should improve (optimizer).
- Fitting a model — letting the model try to find patterns in the data (how does
Xget toy).
Let’s see these in action using the Keras Sequential API to build a model for our regression data. And then we’ll step through each.
Note: If you’re using TensorFlow 2.7.0+, the
fit()function no longer upscales input data to go from(batch_size, )to(batch_size, 1). To fix this, you’ll need to expand the dimension of input data usingtf.expand_dims(input_data, axis=-1).In our case, this means instead of using
model.fit(X, y, epochs=5), usemodel.fit(tf.expand_dims(X, axis=-1), y, epochs=5).
In [25]:
# Set random seed tf.random.set_seed(42) # Create a model using the Sequential API model = tf.keras.Sequential([ tf.keras.layers.Dense(1) ]) # Compile the model model.compile(loss=tf.keras.losses.mae, # mae is short for mean absolute error optimizer=tf.keras.optimizers.SGD(), # SGD is short for stochastic gradient descent metrics=["mae"]) # Fit the model # model.fit(X, y, epochs=5) # this will break with TensorFlow 2.7.0+ model.fit(tf.expand_dims(X, axis=-1), y, epochs=5)
# Set random seed
tf.random.set_seed(42)
# Create a model using the Sequential API
model = tf.keras.Sequential([
tf.keras.layers.Dense(1)
])
# Compile the model
model.compile(loss=tf.keras.losses.mae, # mae is short for mean absolute error
optimizer=tf.keras.optimizers.SGD(), # SGD is short for stochastic gradient descent
metrics=[«mae»])
# Fit the model
# model.fit(X, y, epochs=5) # this will break with TensorFlow 2.7.0+
model.fit(tf.expand_dims(X, axis=-1), y, epochs=5)
Epoch 1/5 1/1 [==============================] - 0s 313ms/step - loss: 11.5048 - mae: 11.5048 Epoch 2/5 1/1 [==============================] - 0s 7ms/step - loss: 11.3723 - mae: 11.3723 Epoch 3/5 1/1 [==============================] - 0s 5ms/step - loss: 11.2398 - mae: 11.2398 Epoch 4/5 1/1 [==============================] - 0s 8ms/step - loss: 11.1073 - mae: 11.1073 Epoch 5/5 1/1 [==============================] - 0s 7ms/step - loss: 10.9748 - mae: 10.9748
Out[25]:
<keras.callbacks.History at 0x7f8df6701950>
Boom!
We’ve just trained a model to figure out the patterns between X and y.
How do you think it went?
Out[26]:
(<tf.Tensor: shape=(8,), dtype=float32, numpy=array([-7., -4., -1., 2., 5., 8., 11., 14.], dtype=float32)>, <tf.Tensor: shape=(8,), dtype=float32, numpy=array([ 3., 6., 9., 12., 15., 18., 21., 24.], dtype=float32)>)
What do you think the outcome should be if we passed our model an X value of 17.0?
In [27]:
# Make a prediction with the model model.predict([17.0])
# Make a prediction with the model
model.predict([17.0])
Out[27]:
array([[12.716021]], dtype=float32)
It doesn’t go very well… it should’ve output something close to 27.0.
🤔 Question: What’s Keras? I thought we were working with TensorFlow but every time we write TensorFlow code,
kerascomes aftertf(e.g.tf.keras.layers.Dense())?
Before TensorFlow 2.0+, Keras was an API designed to be able to build deep learning models with ease. Since TensorFlow 2.0+, its functionality has been tightly integrated within the TensorFlow library.
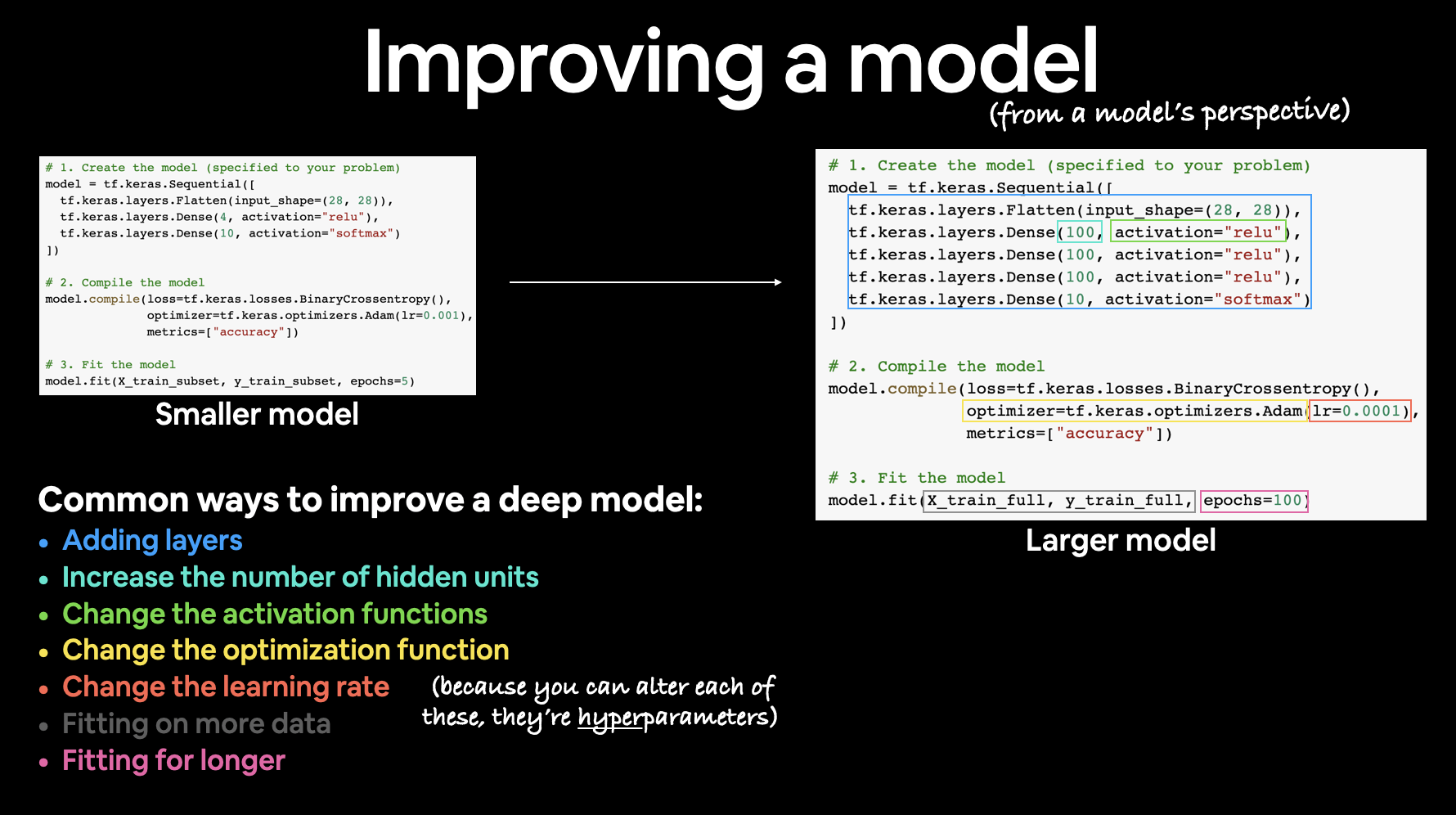
Improving a model¶
How do you think you’d improve upon our current model?
If you guessed by tweaking some of the things we did above, you’d be correct.
To improve our model, we alter almost every part of the 3 steps we went through before.
- Creating a model — here you might want to add more layers, increase the number of hidden units (also called neurons) within each layer, change the activation functions of each layer.
- Compiling a model — you might want to choose optimization function or perhaps change the learning rate of the optimization function.
- Fitting a model — perhaps you could fit a model for more epochs (leave it training for longer) or on more data (give the model more examples to learn from).
There are many different ways to potentially improve a neural network. Some of the most common include: increasing the number of layers (making the network deeper), increasing the number of hidden units (making the network wider) and changing the learning rate. Because these values are all human-changeable, they’re referred to as hyperparameters) and the practice of trying to find the best hyperparameters is referred to as hyperparameter tuning.
Woah. We just introduced a bunch of possible steps. The important thing to remember is how you alter each of these will depend on the problem you’re working on.
And the good thing is, over the next few problems, we’ll get hands-on with all of them.
For now, let’s keep it simple, all we’ll do is train our model for longer (everything else will stay the same).
In [28]:
# Set random seed tf.random.set_seed(42) # Create a model (same as above) model = tf.keras.Sequential([ tf.keras.layers.Dense(1) ]) # Compile model (same as above) model.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=["mae"]) # Fit model (this time we'll train for longer) model.fit(tf.expand_dims(X, axis=-1), y, epochs=100) # train for 100 epochs not 10
# Set random seed
tf.random.set_seed(42)
# Create a model (same as above)
model = tf.keras.Sequential([
tf.keras.layers.Dense(1)
])
# Compile model (same as above)
model.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[«mae»])
# Fit model (this time we’ll train for longer)
model.fit(tf.expand_dims(X, axis=-1), y, epochs=100) # train for 100 epochs not 10
Epoch 1/100 1/1 [==============================] - 0s 319ms/step - loss: 11.5048 - mae: 11.5048 Epoch 2/100 1/1 [==============================] - 0s 6ms/step - loss: 11.3723 - mae: 11.3723 Epoch 3/100 1/1 [==============================] - 0s 5ms/step - loss: 11.2398 - mae: 11.2398 Epoch 4/100 1/1 [==============================] - 0s 4ms/step - loss: 11.1073 - mae: 11.1073 Epoch 5/100 1/1 [==============================] - 0s 6ms/step - loss: 10.9748 - mae: 10.9748 Epoch 6/100 1/1 [==============================] - 0s 6ms/step - loss: 10.8423 - mae: 10.8423 Epoch 7/100 1/1 [==============================] - 0s 6ms/step - loss: 10.7098 - mae: 10.7098 Epoch 8/100 1/1 [==============================] - 0s 9ms/step - loss: 10.5773 - mae: 10.5773 Epoch 9/100 1/1 [==============================] - 0s 7ms/step - loss: 10.4448 - mae: 10.4448 Epoch 10/100 1/1 [==============================] - 0s 6ms/step - loss: 10.3123 - mae: 10.3123 Epoch 11/100 1/1 [==============================] - 0s 14ms/step - loss: 10.1798 - mae: 10.1798 Epoch 12/100 1/1 [==============================] - 0s 9ms/step - loss: 10.0473 - mae: 10.0473 Epoch 13/100 1/1 [==============================] - 0s 7ms/step - loss: 9.9148 - mae: 9.9148 Epoch 14/100 1/1 [==============================] - 0s 8ms/step - loss: 9.7823 - mae: 9.7823 Epoch 15/100 1/1 [==============================] - 0s 9ms/step - loss: 9.6498 - mae: 9.6498 Epoch 16/100 1/1 [==============================] - 0s 5ms/step - loss: 9.5173 - mae: 9.5173 Epoch 17/100 1/1 [==============================] - 0s 6ms/step - loss: 9.3848 - mae: 9.3848 Epoch 18/100 1/1 [==============================] - 0s 6ms/step - loss: 9.2523 - mae: 9.2523 Epoch 19/100 1/1 [==============================] - 0s 5ms/step - loss: 9.1198 - mae: 9.1198 Epoch 20/100 1/1 [==============================] - 0s 5ms/step - loss: 8.9873 - mae: 8.9873 Epoch 21/100 1/1 [==============================] - 0s 6ms/step - loss: 8.8548 - mae: 8.8548 Epoch 22/100 1/1 [==============================] - 0s 6ms/step - loss: 8.7223 - mae: 8.7223 Epoch 23/100 1/1 [==============================] - 0s 7ms/step - loss: 8.5898 - mae: 8.5898 Epoch 24/100 1/1 [==============================] - 0s 4ms/step - loss: 8.4573 - mae: 8.4573 Epoch 25/100 1/1 [==============================] - 0s 6ms/step - loss: 8.3248 - mae: 8.3248 Epoch 26/100 1/1 [==============================] - 0s 7ms/step - loss: 8.1923 - mae: 8.1923 Epoch 27/100 1/1 [==============================] - 0s 5ms/step - loss: 8.0598 - mae: 8.0598 Epoch 28/100 1/1 [==============================] - 0s 5ms/step - loss: 7.9273 - mae: 7.9273 Epoch 29/100 1/1 [==============================] - 0s 5ms/step - loss: 7.7948 - mae: 7.7948 Epoch 30/100 1/1 [==============================] - 0s 5ms/step - loss: 7.6623 - mae: 7.6623 Epoch 31/100 1/1 [==============================] - 0s 7ms/step - loss: 7.5298 - mae: 7.5298 Epoch 32/100 1/1 [==============================] - 0s 4ms/step - loss: 7.3973 - mae: 7.3973 Epoch 33/100 1/1 [==============================] - 0s 5ms/step - loss: 7.2648 - mae: 7.2648 Epoch 34/100 1/1 [==============================] - 0s 6ms/step - loss: 7.2525 - mae: 7.2525 Epoch 35/100 1/1 [==============================] - 0s 7ms/step - loss: 7.2469 - mae: 7.2469 Epoch 36/100 1/1 [==============================] - 0s 5ms/step - loss: 7.2413 - mae: 7.2413 Epoch 37/100 1/1 [==============================] - 0s 5ms/step - loss: 7.2356 - mae: 7.2356 Epoch 38/100 1/1 [==============================] - 0s 6ms/step - loss: 7.2300 - mae: 7.2300 Epoch 39/100 1/1 [==============================] - 0s 6ms/step - loss: 7.2244 - mae: 7.2244 Epoch 40/100 1/1 [==============================] - 0s 5ms/step - loss: 7.2188 - mae: 7.2188 Epoch 41/100 1/1 [==============================] - 0s 7ms/step - loss: 7.2131 - mae: 7.2131 Epoch 42/100 1/1 [==============================] - 0s 5ms/step - loss: 7.2075 - mae: 7.2075 Epoch 43/100 1/1 [==============================] - 0s 7ms/step - loss: 7.2019 - mae: 7.2019 Epoch 44/100 1/1 [==============================] - 0s 5ms/step - loss: 7.1963 - mae: 7.1963 Epoch 45/100 1/1 [==============================] - 0s 6ms/step - loss: 7.1906 - mae: 7.1906 Epoch 46/100 1/1 [==============================] - 0s 5ms/step - loss: 7.1850 - mae: 7.1850 Epoch 47/100 1/1 [==============================] - 0s 7ms/step - loss: 7.1794 - mae: 7.1794 Epoch 48/100 1/1 [==============================] - 0s 5ms/step - loss: 7.1738 - mae: 7.1738 Epoch 49/100 1/1 [==============================] - 0s 5ms/step - loss: 7.1681 - mae: 7.1681 Epoch 50/100 1/1 [==============================] - 0s 5ms/step - loss: 7.1625 - mae: 7.1625 Epoch 51/100 1/1 [==============================] - 0s 6ms/step - loss: 7.1569 - mae: 7.1569 Epoch 52/100 1/1 [==============================] - 0s 6ms/step - loss: 7.1512 - mae: 7.1512 Epoch 53/100 1/1 [==============================] - 0s 6ms/step - loss: 7.1456 - mae: 7.1456 Epoch 54/100 1/1 [==============================] - 0s 31ms/step - loss: 7.1400 - mae: 7.1400 Epoch 55/100 1/1 [==============================] - 0s 7ms/step - loss: 7.1344 - mae: 7.1344 Epoch 56/100 1/1 [==============================] - 0s 6ms/step - loss: 7.1287 - mae: 7.1287 Epoch 57/100 1/1 [==============================] - 0s 7ms/step - loss: 7.1231 - mae: 7.1231 Epoch 58/100 1/1 [==============================] - 0s 8ms/step - loss: 7.1175 - mae: 7.1175 Epoch 59/100 1/1 [==============================] - 0s 11ms/step - loss: 7.1119 - mae: 7.1119 Epoch 60/100 1/1 [==============================] - 0s 7ms/step - loss: 7.1063 - mae: 7.1063 Epoch 61/100 1/1 [==============================] - 0s 7ms/step - loss: 7.1006 - mae: 7.1006 Epoch 62/100 1/1 [==============================] - 0s 7ms/step - loss: 7.0950 - mae: 7.0950 Epoch 63/100 1/1 [==============================] - 0s 7ms/step - loss: 7.0894 - mae: 7.0894 Epoch 64/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0838 - mae: 7.0838 Epoch 65/100 1/1 [==============================] - 0s 6ms/step - loss: 7.0781 - mae: 7.0781 Epoch 66/100 1/1 [==============================] - 0s 7ms/step - loss: 7.0725 - mae: 7.0725 Epoch 67/100 1/1 [==============================] - 0s 6ms/step - loss: 7.0669 - mae: 7.0669 Epoch 68/100 1/1 [==============================] - 0s 5ms/step - loss: 7.0613 - mae: 7.0613 Epoch 69/100 1/1 [==============================] - 0s 7ms/step - loss: 7.0556 - mae: 7.0556 Epoch 70/100 1/1 [==============================] - 0s 26ms/step - loss: 7.0500 - mae: 7.0500 Epoch 71/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0444 - mae: 7.0444 Epoch 72/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0388 - mae: 7.0388 Epoch 73/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0331 - mae: 7.0331 Epoch 74/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0275 - mae: 7.0275 Epoch 75/100 1/1 [==============================] - 0s 5ms/step - loss: 7.0219 - mae: 7.0219 Epoch 76/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0163 - mae: 7.0163 Epoch 77/100 1/1 [==============================] - 0s 8ms/step - loss: 7.0106 - mae: 7.0106 Epoch 78/100 1/1 [==============================] - 0s 7ms/step - loss: 7.0050 - mae: 7.0050 Epoch 79/100 1/1 [==============================] - 0s 6ms/step - loss: 6.9994 - mae: 6.9994 Epoch 80/100 1/1 [==============================] - 0s 10ms/step - loss: 6.9938 - mae: 6.9938 Epoch 81/100 1/1 [==============================] - 0s 10ms/step - loss: 6.9881 - mae: 6.9881 Epoch 82/100 1/1 [==============================] - 0s 6ms/step - loss: 6.9825 - mae: 6.9825 Epoch 83/100 1/1 [==============================] - 0s 8ms/step - loss: 6.9769 - mae: 6.9769 Epoch 84/100 1/1 [==============================] - 0s 8ms/step - loss: 6.9713 - mae: 6.9713 Epoch 85/100 1/1 [==============================] - 0s 12ms/step - loss: 6.9656 - mae: 6.9656 Epoch 86/100 1/1 [==============================] - 0s 6ms/step - loss: 6.9600 - mae: 6.9600 Epoch 87/100 1/1 [==============================] - 0s 16ms/step - loss: 6.9544 - mae: 6.9544 Epoch 88/100 1/1 [==============================] - 0s 9ms/step - loss: 6.9488 - mae: 6.9488 Epoch 89/100 1/1 [==============================] - 0s 11ms/step - loss: 6.9431 - mae: 6.9431 Epoch 90/100 1/1 [==============================] - 0s 6ms/step - loss: 6.9375 - mae: 6.9375 Epoch 91/100 1/1 [==============================] - 0s 5ms/step - loss: 6.9319 - mae: 6.9319 Epoch 92/100 1/1 [==============================] - 0s 7ms/step - loss: 6.9263 - mae: 6.9263 Epoch 93/100 1/1 [==============================] - 0s 7ms/step - loss: 6.9206 - mae: 6.9206 Epoch 94/100 1/1 [==============================] - 0s 5ms/step - loss: 6.9150 - mae: 6.9150 Epoch 95/100 1/1 [==============================] - 0s 8ms/step - loss: 6.9094 - mae: 6.9094 Epoch 96/100 1/1 [==============================] - 0s 6ms/step - loss: 6.9038 - mae: 6.9038 Epoch 97/100 1/1 [==============================] - 0s 7ms/step - loss: 6.8981 - mae: 6.8981 Epoch 98/100 1/1 [==============================] - 0s 10ms/step - loss: 6.8925 - mae: 6.8925 Epoch 99/100 1/1 [==============================] - 0s 5ms/step - loss: 6.8869 - mae: 6.8869 Epoch 100/100 1/1 [==============================] - 0s 13ms/step - loss: 6.8813 - mae: 6.8813
Out[28]:
<keras.callbacks.History at 0x7f8df664db90>
You might’ve noticed the loss value decrease from before (and keep decreasing as the number of epochs gets higher).
What do you think this means for when we make a prediction with our model?
How about we try predict on 17.0 again?
In [29]:
# Remind ourselves of what X and y are X, y
# Remind ourselves of what X and y are
X, y
Out[29]:
(<tf.Tensor: shape=(8,), dtype=float32, numpy=array([-7., -4., -1., 2., 5., 8., 11., 14.], dtype=float32)>, <tf.Tensor: shape=(8,), dtype=float32, numpy=array([ 3., 6., 9., 12., 15., 18., 21., 24.], dtype=float32)>)
In [30]:
# Try and predict what y would be if X was 17.0 model.predict([17.0]) # the right answer is 27.0 (y = X + 10)
# Try and predict what y would be if X was 17.0
model.predict([17.0]) # the right answer is 27.0 (y = X + 10)
Out[30]:
array([[30.158512]], dtype=float32)
Much better!
We got closer this time. But we could still be better.
Now we’ve trained a model, how could we evaluate it?
Evaluating a model¶
A typical workflow you’ll go through when building neural networks is:
Build a model -> evaluate it -> build (tweak) a model -> evaulate it -> build (tweak) a model -> evaluate it...The tweaking comes from maybe not building a model from scratch but adjusting an existing one.
Visualize, visualize, visualize¶
When it comes to evaluation, you’ll want to remember the words: «visualize, visualize, visualize.»
This is because you’re probably better looking at something (doing) than you are thinking about something.
It’s a good idea to visualize:
- The data — what data are you working with? What does it look like?
- The model itself — what does the architecture look like? What are the different shapes?
- The training of a model — how does a model perform while it learns?
- The predictions of a model — how do the predictions of a model line up against the ground truth (the original labels)?
Let’s start by visualizing the model.
But first, we’ll create a little bit of a bigger dataset and a new model we can use (it’ll be the same as before, but the more practice the better).
In [31]:
# Make a bigger dataset X = np.arange(-100, 100, 4) X
# Make a bigger dataset
X = np.arange(-100, 100, 4)
X
Out[31]:
array([-100, -96, -92, -88, -84, -80, -76, -72, -68, -64, -60,
-56, -52, -48, -44, -40, -36, -32, -28, -24, -20, -16,
-12, -8, -4, 0, 4, 8, 12, 16, 20, 24, 28,
32, 36, 40, 44, 48, 52, 56, 60, 64, 68, 72,
76, 80, 84, 88, 92, 96])
In [32]:
# Make labels for the dataset (adhering to the same pattern as before) y = np.arange(-90, 110, 4) y
# Make labels for the dataset (adhering to the same pattern as before)
y = np.arange(-90, 110, 4)
y
Out[32]:
array([-90, -86, -82, -78, -74, -70, -66, -62, -58, -54, -50, -46, -42,
-38, -34, -30, -26, -22, -18, -14, -10, -6, -2, 2, 6, 10,
14, 18, 22, 26, 30, 34, 38, 42, 46, 50, 54, 58, 62,
66, 70, 74, 78, 82, 86, 90, 94, 98, 102, 106])
Since $y=X+10$, we could make the labels like so:
In [33]:
# Same result as above y = X + 10 y
# Same result as above
y = X + 10
y
Out[33]:
array([-90, -86, -82, -78, -74, -70, -66, -62, -58, -54, -50, -46, -42,
-38, -34, -30, -26, -22, -18, -14, -10, -6, -2, 2, 6, 10,
14, 18, 22, 26, 30, 34, 38, 42, 46, 50, 54, 58, 62,
66, 70, 74, 78, 82, 86, 90, 94, 98, 102, 106])
Split data into training/test set¶
One of the other most common and important steps in a machine learning project is creating a training and test set (and when required, a validation set).
Each set serves a specific purpose:
- Training set — the model learns from this data, which is typically 70-80% of the total data available (like the course materials you study during the semester).
- Validation set — the model gets tuned on this data, which is typically 10-15% of the total data available (like the practice exam you take before the final exam).
- Test set — the model gets evaluated on this data to test what it has learned, it’s typically 10-15% of the total data available (like the final exam you take at the end of the semester).
For now, we’ll just use a training and test set, this means we’ll have a dataset for our model to learn on as well as be evaluated on.
We can create them by splitting our X and y arrays.
🔑 Note: When dealing with real-world data, this step is typically done right at the start of a project (the test set should always be kept separate from all other data). We want our model to learn on training data and then evaluate it on test data to get an indication of how well it generalizes to unseen examples.
In [34]:
# Check how many samples we have len(X)
# Check how many samples we have
len(X)
In [35]:
# Split data into train and test sets X_train = X[:40] # first 40 examples (80% of data) y_train = y[:40] X_test = X[40:] # last 10 examples (20% of data) y_test = y[40:] len(X_train), len(X_test)
# Split data into train and test sets
X_train = X[:40] # first 40 examples (80% of data)
y_train = y[:40]
X_test = X[40:] # last 10 examples (20% of data)
y_test = y[40:]
len(X_train), len(X_test)
Visualizing the data¶
Now we’ve got our training and test data, it’s a good idea to visualize it.
Let’s plot it with some nice colours to differentiate what’s what.
In [36]:
plt.figure(figsize=(10, 7)) # Plot training data in blue plt.scatter(X_train, y_train, c='b', label='Training data') # Plot test data in green plt.scatter(X_test, y_test, c='g', label='Testing data') # Show the legend plt.legend();
plt.figure(figsize=(10, 7))
# Plot training data in blue
plt.scatter(X_train, y_train, c=’b’, label=’Training data’)
# Plot test data in green
plt.scatter(X_test, y_test, c=’g’, label=’Testing data’)
# Show the legend
plt.legend();
Beautiful! Any time you can visualize your data, your model, your anything, it’s a good idea.
With this graph in mind, what we’ll be trying to do is build a model which learns the pattern in the blue dots (X_train) to draw the green dots (X_test).
Time to build a model. We’ll make the exact same one from before (the one we trained for longer).
In [37]:
# Set random seed tf.random.set_seed(42) # Create a model (same as above) model = tf.keras.Sequential([ tf.keras.layers.Dense(1) ]) # Compile model (same as above) model.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=["mae"]) # Fit model (same as above) #model.fit(X_train, y_train, epochs=100) # commented out on purpose (not fitting it just yet)
# Set random seed
tf.random.set_seed(42)
# Create a model (same as above)
model = tf.keras.Sequential([
tf.keras.layers.Dense(1)
])
# Compile model (same as above)
model.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[«mae»])
# Fit model (same as above)
#model.fit(X_train, y_train, epochs=100) # commented out on purpose (not fitting it just yet)
Visualizing the model¶
After you’ve built a model, you might want to take a look at it (especially if you haven’t built many before).
You can take a look at the layers and shapes of your model by calling summary() on it.
🔑 Note: Visualizing a model is particularly helpful when you run into input and output shape mismatches.
In [38]:
# Doesn't work (model not fit/built) model.summary()
# Doesn’t work (model not fit/built)
model.summary()
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) <ipython-input-38-7d09d31d4e66> in <module>() 1 # Doesn't work (model not fit/built) ----> 2 model.summary() /usr/local/lib/python3.7/dist-packages/keras/engine/training.py in summary(self, line_length, positions, print_fn, expand_nested) 2578 if not self.built: 2579 raise ValueError( -> 2580 'This model has not yet been built. ' 2581 'Build the model first by calling `build()` or by calling ' 2582 'the model on a batch of data.') ValueError: This model has not yet been built. Build the model first by calling `build()` or by calling the model on a batch of data.
Ahh, the cell above errors because we haven’t fit our built our model.
We also haven’t told it what input shape it should be expecting.
Remember above, how we discussed the input shape was just one number?
We can let our model know the input shape of our data using the input_shape parameter to the first layer (usually if input_shape isn’t defined, Keras tries to figure it out automatically).
In [39]:
# Set random seed tf.random.set_seed(42) # Create a model (same as above) model = tf.keras.Sequential([ tf.keras.layers.Dense(1, input_shape=[1]) # define the input_shape to our model ]) # Compile model (same as above) model.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=["mae"])
# Set random seed
tf.random.set_seed(42)
# Create a model (same as above)
model = tf.keras.Sequential([
tf.keras.layers.Dense(1, input_shape=[1]) # define the input_shape to our model
])
# Compile model (same as above)
model.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[«mae»])
In [40]:
# This will work after specifying the input shape model.summary()
# This will work after specifying the input shape
model.summary()
Model: "sequential_8"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_8 (Dense) (None, 1) 2
=================================================================
Total params: 2
Trainable params: 2
Non-trainable params: 0
_________________________________________________________________
Calling summary() on our model shows us the layers it contains, the output shape and the number of parameters.
- Total params — total number of parameters in the model.
- Trainable parameters — these are the parameters (patterns) the model can update as it trains.
- Non-trainable parameters — these parameters aren’t updated during training (this is typical when you bring in the already learned patterns from other models during transfer learning).
📖 Resource: For a more in-depth overview of the trainable parameters within a layer, check out MIT’s introduction to deep learning video.
🛠 Exercise: Try playing around with the number of hidden units in the
Denselayer (e.g.Dense(2),Dense(3)). How does this change the Total/Trainable params? Investigate what’s causing the change.
For now, all you need to think about these parameters is that their learnable patterns in the data.
Let’s fit our model to the training data.
In [41]:
# Fit the model to the training data model.fit(X_train, y_train, epochs=100, verbose=0) # verbose controls how much gets output
# Fit the model to the training data
model.fit(X_train, y_train, epochs=100, verbose=0) # verbose controls how much gets output
Out[41]:
<keras.callbacks.History at 0x7f8df64e8250>
In [42]:
# Check the model summary model.summary()
# Check the model summary
model.summary()
Model: "sequential_8"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_8 (Dense) (None, 1) 2
=================================================================
Total params: 2
Trainable params: 2
Non-trainable params: 0
_________________________________________________________________
Alongside summary, you can also view a 2D plot of the model using plot_model().
In [43]:
from tensorflow.keras.utils import plot_model plot_model(model, show_shapes=True)
from tensorflow.keras.utils import plot_model
plot_model(model, show_shapes=True)
Out[43]:
In our case, the model we used only has an input and an output but visualizing more complicated models can be very helpful for debugging.
Visualizing the predictions¶
Now we’ve got a trained model, let’s visualize some predictions.
To visualize predictions, it’s always a good idea to plot them against the ground truth labels.
Often you’ll see this in the form of y_test vs. y_pred (ground truth vs. predictions).
First, we’ll make some predictions on the test data (X_test), remember the model has never seen the test data.
In [44]:
# Make predictions y_preds = model.predict(X_test)
# Make predictions
y_preds = model.predict(X_test)
WARNING:tensorflow:5 out of the last 5 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7f8df63c6290> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
In [45]:
# View the predictions y_preds
# View the predictions
y_preds
Out[45]:
array([[53.57109 ],
[57.05633 ],
[60.541573],
[64.02681 ],
[67.512054],
[70.99729 ],
[74.48254 ],
[77.96777 ],
[81.45301 ],
[84.938255]], dtype=float32)
Okay, we get a list of numbers but how do these compare to the ground truth labels?
Let’s build a plotting function to find out.
🔑 Note: If you think you’re going to be visualizing something a lot, it’s a good idea to functionize it so you can use it later.
In [46]:
def plot_predictions(train_data=X_train, train_labels=y_train, test_data=X_test, test_labels=y_test, predictions=y_preds): """ Plots training data, test data and compares predictions. """ plt.figure(figsize=(10, 7)) # Plot training data in blue plt.scatter(train_data, train_labels, c="b", label="Training data") # Plot test data in green plt.scatter(test_data, test_labels, c="g", label="Testing data") # Plot the predictions in red (predictions were made on the test data) plt.scatter(test_data, predictions, c="r", label="Predictions") # Show the legend plt.legend();
def plot_predictions(train_data=X_train,
train_labels=y_train,
test_data=X_test,
test_labels=y_test,
predictions=y_preds):
«»»
Plots training data, test data and compares predictions.
«»»
plt.figure(figsize=(10, 7))
# Plot training data in blue
plt.scatter(train_data, train_labels, c=»b», label=»Training data»)
# Plot test data in green
plt.scatter(test_data, test_labels, c=»g», label=»Testing data»)
# Plot the predictions in red (predictions were made on the test data)
plt.scatter(test_data, predictions, c=»r», label=»Predictions»)
# Show the legend
plt.legend();
In [47]:
plot_predictions(train_data=X_train, train_labels=y_train, test_data=X_test, test_labels=y_test, predictions=y_preds)
plot_predictions(train_data=X_train,
train_labels=y_train,
test_data=X_test,
test_labels=y_test,
predictions=y_preds)
From the plot we can see our predictions aren’t totally outlandish but they definitely aren’t anything special either.
Evaluating predictions¶
Alongisde visualizations, evaulation metrics are your alternative best option for evaluating your model.
Depending on the problem you’re working on, different models have different evaluation metrics.
Two of the main metrics used for regression problems are:
- Mean absolute error (MAE) — the mean difference between each of the predictions.
- Mean squared error (MSE) — the squared mean difference between of the predictions (use if larger errors are more detrimental than smaller errors).
The lower each of these values, the better.
You can also use model.evaluate() which will return the loss of the model as well as any metrics setup during the compile step.
In [48]:
# Evaluate the model on the test set model.evaluate(X_test, y_test)
# Evaluate the model on the test set
model.evaluate(X_test, y_test)
1/1 [==============================] - 0s 137ms/step - loss: 18.7453 - mae: 18.7453
Out[48]:
[18.74532699584961, 18.74532699584961]
In our case, since we used MAE for the loss function as well as MAE for the metrics, model.evaulate() returns them both.
TensorFlow also has built in functions for MSE and MAE.
For many evaluation functions, the premise is the same: compare predictions to the ground truth labels.
In [49]:
# Calculate the mean absolute error mae = tf.metrics.mean_absolute_error(y_true=y_test, y_pred=y_preds) mae
# Calculate the mean absolute error
mae = tf.metrics.mean_absolute_error(y_true=y_test,
y_pred=y_preds)
mae
Out[49]:
<tf.Tensor: shape=(10,), dtype=float32, numpy=
array([34.42891 , 30.943668, 27.45843 , 23.97319 , 20.487946, 17.202168,
14.510478, 12.419336, 11.018796, 10.212349], dtype=float32)>
Huh? That’s strange, MAE should be a single output.
Instead, we get 10 values.
This is because our y_test and y_preds tensors are different shapes.
In [50]:
# Check the test label tensor values y_test
# Check the test label tensor values
y_test
Out[50]:
array([ 70, 74, 78, 82, 86, 90, 94, 98, 102, 106])
In [51]:
# Check the predictions tensor values (notice the extra square brackets) y_preds
# Check the predictions tensor values (notice the extra square brackets)
y_preds
Out[51]:
array([[53.57109 ],
[57.05633 ],
[60.541573],
[64.02681 ],
[67.512054],
[70.99729 ],
[74.48254 ],
[77.96777 ],
[81.45301 ],
[84.938255]], dtype=float32)
In [52]:
# Check the tensor shapes y_test.shape, y_preds.shape
# Check the tensor shapes
y_test.shape, y_preds.shape
Remember how we discussed dealing with different input and output shapes is one the most common issues you’ll come across, this is one of those times.
But not to worry.
We can fix it using squeeze(), it’ll remove the the 1 dimension from our y_preds tensor, making it the same shape as y_test.
🔑 Note: If you’re comparing two tensors, it’s important to make sure they’re the right shape(s) (you won’t always have to manipulate the shapes, but always be on the look out, many errors are the result of mismatched tensors, especially mismatched input and output shapes).
In [53]:
# Shape before squeeze() y_preds.shape
# Shape before squeeze()
y_preds.shape
In [54]:
# Shape after squeeze() y_preds.squeeze().shape
# Shape after squeeze()
y_preds.squeeze().shape
In [55]:
# What do they look like? y_test, y_preds.squeeze()
# What do they look like?
y_test, y_preds.squeeze()
Out[55]:
(array([ 70, 74, 78, 82, 86, 90, 94, 98, 102, 106]),
array([53.57109 , 57.05633 , 60.541573, 64.02681 , 67.512054, 70.99729 ,
74.48254 , 77.96777 , 81.45301 , 84.938255], dtype=float32))
Okay, now we know how to make our y_test and y_preds tenors the same shape, let’s use our evaluation metrics.
In [56]:
# Calcuate the MAE mae = tf.metrics.mean_absolute_error(y_true=y_test, y_pred=y_preds.squeeze()) # use squeeze() to make same shape mae
# Calcuate the MAE
mae = tf.metrics.mean_absolute_error(y_true=y_test,
y_pred=y_preds.squeeze()) # use squeeze() to make same shape
mae
Out[56]:
<tf.Tensor: shape=(), dtype=float32, numpy=18.745327>
In [57]:
# Calculate the MSE mse = tf.metrics.mean_squared_error(y_true=y_test, y_pred=y_preds.squeeze()) mse
# Calculate the MSE
mse = tf.metrics.mean_squared_error(y_true=y_test,
y_pred=y_preds.squeeze())
mse
Out[57]:
<tf.Tensor: shape=(), dtype=float32, numpy=353.57336>
We can also calculate the MAE using pure TensorFlow functions.
In [58]:
# Returns the same as tf.metrics.mean_absolute_error() tf.reduce_mean(tf.abs(y_test-y_preds.squeeze()))
# Returns the same as tf.metrics.mean_absolute_error()
tf.reduce_mean(tf.abs(y_test-y_preds.squeeze()))
Out[58]:
<tf.Tensor: shape=(), dtype=float64, numpy=18.745327377319335>
Again, it’s a good idea to functionize anything you think you might use over again (or find yourself using over and over again).
Let’s make functions for our evaluation metrics.
In [59]:
def mae(y_test, y_pred): """ Calculuates mean absolute error between y_test and y_preds. """ return tf.metrics.mean_absolute_error(y_test, y_pred) def mse(y_test, y_pred): """ Calculates mean squared error between y_test and y_preds. """ return tf.metrics.mean_squared_error(y_test, y_pred)
def mae(y_test, y_pred):
«»»
Calculuates mean absolute error between y_test and y_preds.
«»»
return tf.metrics.mean_absolute_error(y_test,
y_pred)
def mse(y_test, y_pred):
«»»
Calculates mean squared error between y_test and y_preds.
«»»
return tf.metrics.mean_squared_error(y_test,
y_pred)
Running experiments to improve a model¶
After seeing the evaluation metrics and the predictions your model makes, it’s likely you’ll want to improve it.
Again, there are many different ways you can do this, but 3 of the main ones are:
- Get more data — get more examples for your model to train on (more opportunities to learn patterns).
- Make your model larger (use a more complex model) — this might come in the form of more layers or more hidden units in each layer.
- Train for longer — give your model more of a chance to find the patterns in the data.
Since we created our dataset, we could easily make more data but this isn’t always the case when you’re working with real-world datasets.
So let’s take a look at how we can improve our model using 2 and 3.
To do so, we’ll build 3 models and compare their results:
model_1— same as original model, 1 layer, trained for 100 epochs.model_2— 2 layers, trained for 100 epochs.model_3— 2 layers, trained for 500 epochs.
Build model_1
In [63]:
# Set random seed tf.random.set_seed(42) # Replicate original model model_1 = tf.keras.Sequential([ tf.keras.layers.Dense(1) ]) # Compile the model model_1.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=['mae']) # Fit the model model_1.fit(tf.expand_dims(X_train, axis=-1), y_train, epochs=100)
# Set random seed
tf.random.set_seed(42)
# Replicate original model
model_1 = tf.keras.Sequential([
tf.keras.layers.Dense(1)
])
# Compile the model
model_1.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[‘mae’])
# Fit the model
model_1.fit(tf.expand_dims(X_train, axis=-1), y_train, epochs=100)
Epoch 1/100 2/2 [==============================] - 0s 9ms/step - loss: 15.9024 - mae: 15.9024 Epoch 2/100 2/2 [==============================] - 0s 5ms/step - loss: 11.2837 - mae: 11.2837 Epoch 3/100 2/2 [==============================] - 0s 5ms/step - loss: 11.1074 - mae: 11.1074 Epoch 4/100 2/2 [==============================] - 0s 7ms/step - loss: 9.2991 - mae: 9.2991 Epoch 5/100 2/2 [==============================] - 0s 5ms/step - loss: 10.1677 - mae: 10.1677 Epoch 6/100 2/2 [==============================] - 0s 5ms/step - loss: 9.4303 - mae: 9.4303 Epoch 7/100 2/2 [==============================] - 0s 8ms/step - loss: 8.5704 - mae: 8.5704 Epoch 8/100 2/2 [==============================] - 0s 4ms/step - loss: 9.0442 - mae: 9.0442 Epoch 9/100 2/2 [==============================] - 0s 6ms/step - loss: 18.7517 - mae: 18.7517 Epoch 10/100 2/2 [==============================] - 0s 4ms/step - loss: 10.1142 - mae: 10.1142 Epoch 11/100 2/2 [==============================] - 0s 8ms/step - loss: 8.3980 - mae: 8.3980 Epoch 12/100 2/2 [==============================] - 0s 5ms/step - loss: 10.6639 - mae: 10.6639 Epoch 13/100 2/2 [==============================] - 0s 5ms/step - loss: 9.7977 - mae: 9.7977 Epoch 14/100 2/2 [==============================] - 0s 16ms/step - loss: 16.0103 - mae: 16.0103 Epoch 15/100 2/2 [==============================] - 0s 9ms/step - loss: 11.4068 - mae: 11.4068 Epoch 16/100 2/2 [==============================] - 0s 4ms/step - loss: 8.5393 - mae: 8.5393 Epoch 17/100 2/2 [==============================] - 0s 7ms/step - loss: 13.6348 - mae: 13.6348 Epoch 18/100 2/2 [==============================] - 0s 7ms/step - loss: 11.4629 - mae: 11.4629 Epoch 19/100 2/2 [==============================] - 0s 10ms/step - loss: 17.9148 - mae: 17.9148 Epoch 20/100 2/2 [==============================] - 0s 6ms/step - loss: 15.0494 - mae: 15.0494 Epoch 21/100 2/2 [==============================] - 0s 6ms/step - loss: 11.0216 - mae: 11.0216 Epoch 22/100 2/2 [==============================] - 0s 7ms/step - loss: 8.1558 - mae: 8.1558 Epoch 23/100 2/2 [==============================] - 0s 9ms/step - loss: 9.5138 - mae: 9.5138 Epoch 24/100 2/2 [==============================] - 0s 18ms/step - loss: 7.6617 - mae: 7.6617 Epoch 25/100 2/2 [==============================] - 0s 10ms/step - loss: 13.1859 - mae: 13.1859 Epoch 26/100 2/2 [==============================] - 0s 6ms/step - loss: 16.4211 - mae: 16.4211 Epoch 27/100 2/2 [==============================] - 0s 6ms/step - loss: 13.1660 - mae: 13.1660 Epoch 28/100 2/2 [==============================] - 0s 5ms/step - loss: 14.2559 - mae: 14.2559 Epoch 29/100 2/2 [==============================] - 0s 6ms/step - loss: 10.0670 - mae: 10.0670 Epoch 30/100 2/2 [==============================] - 0s 9ms/step - loss: 16.3409 - mae: 16.3409 Epoch 31/100 2/2 [==============================] - 0s 4ms/step - loss: 23.6444 - mae: 23.6444 Epoch 32/100 2/2 [==============================] - 0s 4ms/step - loss: 7.6215 - mae: 7.6215 Epoch 33/100 2/2 [==============================] - 0s 11ms/step - loss: 9.3221 - mae: 9.3221 Epoch 34/100 2/2 [==============================] - 0s 6ms/step - loss: 13.7313 - mae: 13.7313 Epoch 35/100 2/2 [==============================] - 0s 5ms/step - loss: 11.1276 - mae: 11.1276 Epoch 36/100 2/2 [==============================] - 0s 4ms/step - loss: 13.3222 - mae: 13.3222 Epoch 37/100 2/2 [==============================] - 0s 5ms/step - loss: 9.4763 - mae: 9.4763 Epoch 38/100 2/2 [==============================] - 0s 5ms/step - loss: 10.1381 - mae: 10.1381 Epoch 39/100 2/2 [==============================] - 0s 5ms/step - loss: 10.1793 - mae: 10.1793 Epoch 40/100 2/2 [==============================] - 0s 4ms/step - loss: 10.9137 - mae: 10.9137 Epoch 41/100 2/2 [==============================] - 0s 5ms/step - loss: 7.9063 - mae: 7.9063 Epoch 42/100 2/2 [==============================] - 0s 4ms/step - loss: 10.0914 - mae: 10.0914 Epoch 43/100 2/2 [==============================] - 0s 5ms/step - loss: 8.7006 - mae: 8.7006 Epoch 44/100 2/2 [==============================] - 0s 3ms/step - loss: 12.2047 - mae: 12.2047 Epoch 45/100 2/2 [==============================] - 0s 4ms/step - loss: 13.7970 - mae: 13.7970 Epoch 46/100 2/2 [==============================] - 0s 4ms/step - loss: 8.4687 - mae: 8.4687 Epoch 47/100 2/2 [==============================] - 0s 5ms/step - loss: 9.1330 - mae: 9.1330 Epoch 48/100 2/2 [==============================] - 0s 4ms/step - loss: 10.6190 - mae: 10.6190 Epoch 49/100 2/2 [==============================] - 0s 4ms/step - loss: 7.7503 - mae: 7.7503 Epoch 50/100 2/2 [==============================] - 0s 4ms/step - loss: 9.5407 - mae: 9.5407 Epoch 51/100 2/2 [==============================] - 0s 4ms/step - loss: 9.1584 - mae: 9.1584 Epoch 52/100 2/2 [==============================] - 0s 4ms/step - loss: 16.3630 - mae: 16.3630 Epoch 53/100 2/2 [==============================] - 0s 4ms/step - loss: 14.1299 - mae: 14.1299 Epoch 54/100 2/2 [==============================] - 0s 4ms/step - loss: 21.1247 - mae: 21.1247 Epoch 55/100 2/2 [==============================] - 0s 5ms/step - loss: 16.3961 - mae: 16.3961 Epoch 56/100 2/2 [==============================] - 0s 5ms/step - loss: 9.9806 - mae: 9.9806 Epoch 57/100 2/2 [==============================] - 0s 4ms/step - loss: 9.9606 - mae: 9.9606 Epoch 58/100 2/2 [==============================] - 0s 4ms/step - loss: 9.2209 - mae: 9.2209 Epoch 59/100 2/2 [==============================] - 0s 5ms/step - loss: 8.4239 - mae: 8.4239 Epoch 60/100 2/2 [==============================] - 0s 7ms/step - loss: 9.4869 - mae: 9.4869 Epoch 61/100 2/2 [==============================] - 0s 4ms/step - loss: 11.4355 - mae: 11.4355 Epoch 62/100 2/2 [==============================] - 0s 4ms/step - loss: 11.6887 - mae: 11.6887 Epoch 63/100 2/2 [==============================] - 0s 3ms/step - loss: 7.0838 - mae: 7.0838 Epoch 64/100 2/2 [==============================] - 0s 4ms/step - loss: 16.9675 - mae: 16.9675 Epoch 65/100 2/2 [==============================] - 0s 4ms/step - loss: 12.4599 - mae: 12.4599 Epoch 66/100 2/2 [==============================] - 0s 4ms/step - loss: 13.0184 - mae: 13.0184 Epoch 67/100 2/2 [==============================] - 0s 5ms/step - loss: 8.0600 - mae: 8.0600 Epoch 68/100 2/2 [==============================] - 0s 5ms/step - loss: 10.1888 - mae: 10.1888 Epoch 69/100 2/2 [==============================] - 0s 5ms/step - loss: 12.3633 - mae: 12.3633 Epoch 70/100 2/2 [==============================] - 0s 10ms/step - loss: 9.0516 - mae: 9.0516 Epoch 71/100 2/2 [==============================] - 0s 11ms/step - loss: 10.0378 - mae: 10.0378 Epoch 72/100 2/2 [==============================] - 0s 9ms/step - loss: 10.0516 - mae: 10.0516 Epoch 73/100 2/2 [==============================] - 0s 5ms/step - loss: 12.6151 - mae: 12.6151 Epoch 74/100 2/2 [==============================] - 0s 4ms/step - loss: 10.3819 - mae: 10.3819 Epoch 75/100 2/2 [==============================] - 0s 4ms/step - loss: 9.7229 - mae: 9.7229 Epoch 76/100 2/2 [==============================] - 0s 5ms/step - loss: 11.2252 - mae: 11.2252 Epoch 77/100 2/2 [==============================] - 0s 4ms/step - loss: 8.3642 - mae: 8.3642 Epoch 78/100 2/2 [==============================] - 0s 4ms/step - loss: 9.1274 - mae: 9.1274 Epoch 79/100 2/2 [==============================] - 0s 4ms/step - loss: 19.5039 - mae: 19.5039 Epoch 80/100 2/2 [==============================] - 0s 4ms/step - loss: 14.8945 - mae: 14.8945 Epoch 81/100 2/2 [==============================] - 0s 4ms/step - loss: 9.0034 - mae: 9.0034 Epoch 82/100 2/2 [==============================] - 0s 7ms/step - loss: 13.0206 - mae: 13.0206 Epoch 83/100 2/2 [==============================] - 0s 5ms/step - loss: 7.9299 - mae: 7.9299 Epoch 84/100 2/2 [==============================] - 0s 3ms/step - loss: 7.6872 - mae: 7.6872 Epoch 85/100 2/2 [==============================] - 0s 6ms/step - loss: 10.0328 - mae: 10.0328 Epoch 86/100 2/2 [==============================] - 0s 13ms/step - loss: 9.2433 - mae: 9.2433 Epoch 87/100 2/2 [==============================] - 0s 6ms/step - loss: 12.0209 - mae: 12.0209 Epoch 88/100 2/2 [==============================] - 0s 25ms/step - loss: 10.6389 - mae: 10.6389 Epoch 89/100 2/2 [==============================] - 0s 13ms/step - loss: 7.2667 - mae: 7.2667 Epoch 90/100 2/2 [==============================] - 0s 7ms/step - loss: 12.7786 - mae: 12.7786 Epoch 91/100 2/2 [==============================] - 0s 7ms/step - loss: 7.3481 - mae: 7.3481 Epoch 92/100 2/2 [==============================] - 0s 6ms/step - loss: 7.7175 - mae: 7.7175 Epoch 93/100 2/2 [==============================] - 0s 8ms/step - loss: 7.1263 - mae: 7.1263 Epoch 94/100 2/2 [==============================] - 0s 10ms/step - loss: 12.6190 - mae: 12.6190 Epoch 95/100 2/2 [==============================] - 0s 9ms/step - loss: 10.0912 - mae: 10.0912 Epoch 96/100 2/2 [==============================] - 0s 5ms/step - loss: 9.3558 - mae: 9.3558 Epoch 97/100 2/2 [==============================] - 0s 18ms/step - loss: 12.6834 - mae: 12.6834 Epoch 98/100 2/2 [==============================] - 0s 4ms/step - loss: 8.6762 - mae: 8.6762 Epoch 99/100 2/2 [==============================] - 0s 9ms/step - loss: 9.4693 - mae: 9.4693 Epoch 100/100 2/2 [==============================] - 0s 4ms/step - loss: 8.7067 - mae: 8.7067
Out[63]:
<keras.callbacks.History at 0x7f8e751f5a10>
In [64]:
# Make and plot predictions for model_1 y_preds_1 = model_1.predict(X_test) plot_predictions(predictions=y_preds_1)
# Make and plot predictions for model_1
y_preds_1 = model_1.predict(X_test)
plot_predictions(predictions=y_preds_1)
WARNING:tensorflow:6 out of the last 6 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7f8df63b8f80> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
In [65]:
# Calculate model_1 metrics mae_1 = mae(y_test, y_preds_1.squeeze()).numpy() mse_1 = mse(y_test, y_preds_1.squeeze()).numpy() mae_1, mse_1
# Calculate model_1 metrics
mae_1 = mae(y_test, y_preds_1.squeeze()).numpy()
mse_1 = mse(y_test, y_preds_1.squeeze()).numpy()
mae_1, mse_1
Build model_2
This time we’ll add an extra dense layer (so now our model will have 2 layers) whilst keeping everything else the same.
In [67]:
# Set random seed tf.random.set_seed(42) # Replicate model_1 and add an extra layer model_2 = tf.keras.Sequential([ tf.keras.layers.Dense(1), tf.keras.layers.Dense(1) # add a second layer ]) # Compile the model model_2.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=['mae']) # Fit the model model_2.fit(tf.expand_dims(X_train, axis=-1), y_train, epochs=100, verbose=0) # set verbose to 0 for less output
# Set random seed
tf.random.set_seed(42)
# Replicate model_1 and add an extra layer
model_2 = tf.keras.Sequential([
tf.keras.layers.Dense(1),
tf.keras.layers.Dense(1) # add a second layer
])
# Compile the model
model_2.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[‘mae’])
# Fit the model
model_2.fit(tf.expand_dims(X_train, axis=-1), y_train, epochs=100, verbose=0) # set verbose to 0 for less output
Out[67]:
<keras.callbacks.History at 0x7f8df6923cd0>
In [68]:
# Make and plot predictions for model_2 y_preds_2 = model_2.predict(X_test) plot_predictions(predictions=y_preds_2)
# Make and plot predictions for model_2
y_preds_2 = model_2.predict(X_test)
plot_predictions(predictions=y_preds_2)
Woah, that’s looking better already! And all it took was an extra layer.
In [69]:
# Calculate model_2 metrics mae_2 = mae(y_test, y_preds_2.squeeze()).numpy() mse_2 = mse(y_test, y_preds_2.squeeze()).numpy() mae_2, mse_2
# Calculate model_2 metrics
mae_2 = mae(y_test, y_preds_2.squeeze()).numpy()
mse_2 = mse(y_test, y_preds_2.squeeze()).numpy()
mae_2, mse_2
Build model_3
For our 3rd model, we’ll keep everything the same as model_2 except this time we’ll train for longer (500 epochs instead of 100).
This will give our model more of a chance to learn the patterns in the data.
In [71]:
# Set random seed tf.random.set_seed(42) # Replicate model_2 model_3 = tf.keras.Sequential([ tf.keras.layers.Dense(1), tf.keras.layers.Dense(1) ]) # Compile the model model_3.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=['mae']) # Fit the model (this time for 500 epochs, not 100) model_3.fit(tf.expand_dims(X_train, axis=-1), y_train, epochs=500, verbose=0) # set verbose to 0 for less output
# Set random seed
tf.random.set_seed(42)
# Replicate model_2
model_3 = tf.keras.Sequential([
tf.keras.layers.Dense(1),
tf.keras.layers.Dense(1)
])
# Compile the model
model_3.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[‘mae’])
# Fit the model (this time for 500 epochs, not 100)
model_3.fit(tf.expand_dims(X_train, axis=-1), y_train, epochs=500, verbose=0) # set verbose to 0 for less output
Out[71]:
<keras.callbacks.History at 0x7f8df67b6d90>
In [72]:
# Make and plot predictions for model_3 y_preds_3 = model_3.predict(X_test) plot_predictions(predictions=y_preds_3)
# Make and plot predictions for model_3
y_preds_3 = model_3.predict(X_test)
plot_predictions(predictions=y_preds_3)
Strange, we trained for longer but our model performed worse?
As it turns out, our model might’ve trained too long and has thus resulted in worse results (we’ll see ways to prevent training for too long later on).
In [73]:
# Calculate model_3 metrics mae_3 = mae(y_test, y_preds_3.squeeze()).numpy() mse_3 = mse(y_test, y_preds_3.squeeze()).numpy() mae_3, mse_3
# Calculate model_3 metrics
mae_3 = mae(y_test, y_preds_3.squeeze()).numpy()
mse_3 = mse(y_test, y_preds_3.squeeze()).numpy()
mae_3, mse_3
Comparing results¶
Now we’ve got results for 3 similar but slightly different results, let’s compare them.
In [74]:
model_results = [["model_1", mae_1, mse_1], ["model_2", mae_2, mse_2], ["model_3", mae_3, mae_3]]
model_results = [[«model_1», mae_1, mse_1],
[«model_2», mae_2, mse_2],
[«model_3», mae_3, mae_3]]
In [75]:
import pandas as pd all_results = pd.DataFrame(model_results, columns=["model", "mae", "mse"]) all_results
import pandas as pd
all_results = pd.DataFrame(model_results, columns=[«model», «mae», «mse»])
all_results
Out[75]:
| model | mae | mse | |
|---|---|---|---|
| 0 | model_1 | 18.745327 | 353.573364 |
| 1 | model_2 | 1.909811 | 5.459232 |
| 2 | model_3 | 68.687859 | 68.687859 |
From our experiments, it looks like model_2 performed the best.
And now, you might be thinking, «wow, comparing models is tedious…» and it definitely can be, we’ve only compared 3 models here.
But this is part of what machine learning modelling is about, trying many different combinations of models and seeing which performs best.
Each model you build is a small experiment.
🔑 Note: One of your main goals should be to minimize the time between your experiments. The more experiments you do, the more things you’ll figure out which don’t work and in turn, get closer to figuring out what does work. Remember the machine learning practitioner’s motto: «experiment, experiment, experiment».
Another thing you’ll also find is what you thought may work (such as training a model for longer) may not always work and the exact opposite is also often the case.
Tracking your experiments¶
One really good habit to get into is tracking your modelling experiments to see which perform better than others.
We’ve done a simple version of this above (keeping the results in different variables).
📖 Resource: But as you build more models, you’ll want to look into using tools such as:
- TensorBoard — a component of the TensorFlow library to help track modelling experiments (we’ll see this later).
- Weights & Biases — a tool for tracking all kinds of machine learning experiments (the good news for Weights & Biases is it plugs into TensorBoard).
Saving a model¶
Once you’ve trained a model and found one which performs to your liking, you’ll probably want to save it for use elsewhere (like a web application or mobile device).
You can save a TensorFlow/Keras model using model.save().
There are two ways to save a model in TensorFlow:
- The SavedModel format (default).
- The HDF5 format.
The main difference between the two is the SavedModel is automatically able to save custom objects (such as special layers) without additional modifications when loading the model back in.
Which one should you use?
It depends on your situation but the SavedModel format will suffice most of the time.
Both methods use the same method call.
In [76]:
# Save a model using the SavedModel format model_2.save('best_model_SavedModel_format')
# Save a model using the SavedModel format
model_2.save(‘best_model_SavedModel_format’)
INFO:tensorflow:Assets written to: best_model_SavedModel_format/assets
In [77]:
# Check it out - outputs a protobuf binary file (.pb) as well as other files !ls best_model_SavedModel_format
# Check it out — outputs a protobuf binary file (.pb) as well as other files
!ls best_model_SavedModel_format
assets keras_metadata.pb saved_model.pb variables
Now let’s save the model in the HDF5 format, we’ll use the same method but with a different filename.
In [78]:
# Save a model using the HDF5 format model_2.save("best_model_HDF5_format.h5") # note the addition of '.h5' on the end
# Save a model using the HDF5 format
model_2.save(«best_model_HDF5_format.h5») # note the addition of ‘.h5’ on the end
In [79]:
# Check it out !ls best_model_HDF5_format.h5
# Check it out
!ls best_model_HDF5_format.h5
best_model_HDF5_format.h5
Loading a model¶
We can load a saved model using the load_model() method.
Loading a model for the different formats (SavedModel and HDF5) is the same (as long as the pathnames to the particular formats are correct).
In [80]:
# Load a model from the SavedModel format loaded_saved_model = tf.keras.models.load_model("best_model_SavedModel_format") loaded_saved_model.summary()
# Load a model from the SavedModel format
loaded_saved_model = tf.keras.models.load_model(«best_model_SavedModel_format»)
loaded_saved_model.summary()
Model: "sequential_12"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_13 (Dense) (None, 1) 2
dense_14 (Dense) (None, 1) 2
=================================================================
Total params: 4
Trainable params: 4
Non-trainable params: 0
_________________________________________________________________
In [81]:
# Compare model_2 with the SavedModel version (should return True) model_2_preds = model_2.predict(X_test) saved_model_preds = loaded_saved_model.predict(X_test) mae(y_test, saved_model_preds.squeeze()).numpy() == mae(y_test, model_2_preds.squeeze()).numpy()
# Compare model_2 with the SavedModel version (should return True)
model_2_preds = model_2.predict(X_test)
saved_model_preds = loaded_saved_model.predict(X_test)
mae(y_test, saved_model_preds.squeeze()).numpy() == mae(y_test, model_2_preds.squeeze()).numpy()
Loading in from the HDF5 is much the same.
In [82]:
# Load a model from the HDF5 format loaded_h5_model = tf.keras.models.load_model("best_model_HDF5_format.h5") loaded_h5_model.summary()
# Load a model from the HDF5 format
loaded_h5_model = tf.keras.models.load_model(«best_model_HDF5_format.h5»)
loaded_h5_model.summary()
Model: "sequential_12"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_13 (Dense) (None, 1) 2
dense_14 (Dense) (None, 1) 2
=================================================================
Total params: 4
Trainable params: 4
Non-trainable params: 0
_________________________________________________________________
In [83]:
# Compare model_2 with the loaded HDF5 version (should return True) h5_model_preds = loaded_h5_model.predict(X_test) mae(y_test, h5_model_preds.squeeze()).numpy() == mae(y_test, model_2_preds.squeeze()).numpy()
# Compare model_2 with the loaded HDF5 version (should return True)
h5_model_preds = loaded_h5_model.predict(X_test)
mae(y_test, h5_model_preds.squeeze()).numpy() == mae(y_test, model_2_preds.squeeze()).numpy()
Downloading a model (from Google Colab)¶
Say you wanted to get your model from Google Colab to your local machine, you can do one of the following things:
- Right click on the file in the files pane and click ‘download’.
- Use the code below.
In [84]:
# Download the model (or any file) from Google Colab from google.colab import files files.download("best_model_HDF5_format.h5")
# Download the model (or any file) from Google Colab
from google.colab import files
files.download(«best_model_HDF5_format.h5»)
A larger example¶
Alright, we’ve seen the fundamentals of building neural network regression models in TensorFlow.
Let’s step it up a notch and build a model for a more feature rich dataset.
More specifically we’re going to try predict the cost of medical insurance for individuals based on a number of different parameters such as, age, sex, bmi, children, smoking_status and residential_region.
To do, we’ll leverage the pubically available Medical Cost dataset available from Kaggle and hosted on GitHub.
🔑 Note: When learning machine learning paradigms, you’ll often go through a series of foundational techniques and then practice them by working with open-source datasets and examples. Just as we’re doing now, learn foundations, put them to work with different problems. Every time you work on something new, it’s a good idea to search for something like «problem X example with Python/TensorFlow» where you substitute X for your problem.
In [85]:
# Import required libraries import tensorflow as tf import pandas as pd import matplotlib.pyplot as plt
# Import required libraries
import tensorflow as tf
import pandas as pd
import matplotlib.pyplot as plt
In [86]:
# Read in the insurance dataset insurance = pd.read_csv("https://raw.githubusercontent.com/stedy/Machine-Learning-with-R-datasets/master/insurance.csv")
# Read in the insurance dataset
insurance = pd.read_csv(«https://raw.githubusercontent.com/stedy/Machine-Learning-with-R-datasets/master/insurance.csv»)
In [87]:
# Check out the insurance dataset insurance.head()
# Check out the insurance dataset
insurance.head()
Out[87]:
| age | sex | bmi | children | smoker | region | charges | |
|---|---|---|---|---|---|---|---|
| 0 | 19 | female | 27.900 | 0 | yes | southwest | 16884.92400 |
| 1 | 18 | male | 33.770 | 1 | no | southeast | 1725.55230 |
| 2 | 28 | male | 33.000 | 3 | no | southeast | 4449.46200 |
| 3 | 33 | male | 22.705 | 0 | no | northwest | 21984.47061 |
| 4 | 32 | male | 28.880 | 0 | no | northwest | 3866.85520 |
We’re going to have to turn the non-numerical columns into numbers (because a neural network can’t handle non-numerical inputs).
To do so, we’ll use the get_dummies() method in pandas.
It converts categorical variables (like the sex, smoker and region columns) into numerical variables using one-hot encoding.
In [88]:
# Turn all categories into numbers insurance_one_hot = pd.get_dummies(insurance) insurance_one_hot.head() # view the converted columns
# Turn all categories into numbers
insurance_one_hot = pd.get_dummies(insurance)
insurance_one_hot.head() # view the converted columns
Out[88]:
| age | bmi | children | charges | sex_female | sex_male | smoker_no | smoker_yes | region_northeast | region_northwest | region_southeast | region_southwest | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 19 | 27.900 | 0 | 16884.92400 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| 1 | 18 | 33.770 | 1 | 1725.55230 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 2 | 28 | 33.000 | 3 | 4449.46200 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 3 | 33 | 22.705 | 0 | 21984.47061 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 4 | 32 | 28.880 | 0 | 3866.85520 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
Now we’ll split data into features (X) and labels (y).
In [89]:
# Create X & y values X = insurance_one_hot.drop("charges", axis=1) y = insurance_one_hot["charges"]
# Create X & y values
X = insurance_one_hot.drop(«charges», axis=1)
y = insurance_one_hot[«charges»]
Out[90]:
| age | bmi | children | sex_female | sex_male | smoker_no | smoker_yes | region_northeast | region_northwest | region_southeast | region_southwest | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 19 | 27.900 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| 1 | 18 | 33.770 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 2 | 28 | 33.000 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 3 | 33 | 22.705 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 4 | 32 | 28.880 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
And create training and test sets. We could do this manually, but to make it easier, we’ll leverage the already available train_test_split function available from Scikit-Learn.
In [91]:
# Create training and test sets from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42) # set random state for reproducible splits
# Create training and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.2,
random_state=42) # set random state for reproducible splits
Now we can build and fit a model (we’ll make it the same as model_2).
In [92]:
# Set random seed tf.random.set_seed(42) # Create a new model (same as model_2) insurance_model = tf.keras.Sequential([ tf.keras.layers.Dense(1), tf.keras.layers.Dense(1) ]) # Compile the model insurance_model.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.SGD(), metrics=['mae']) # Fit the model insurance_model.fit(X_train, y_train, epochs=100)
# Set random seed
tf.random.set_seed(42)
# Create a new model (same as model_2)
insurance_model = tf.keras.Sequential([
tf.keras.layers.Dense(1),
tf.keras.layers.Dense(1)
])
# Compile the model
insurance_model.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.SGD(),
metrics=[‘mae’])
# Fit the model
insurance_model.fit(X_train, y_train, epochs=100)
Epoch 1/100 34/34 [==============================] - 0s 1ms/step - loss: 8868.5918 - mae: 8868.5918 Epoch 2/100 34/34 [==============================] - 0s 1ms/step - loss: 7887.1606 - mae: 7887.1606 Epoch 3/100 34/34 [==============================] - 0s 1ms/step - loss: 7537.0947 - mae: 7537.0947 Epoch 4/100 34/34 [==============================] - 0s 1ms/step - loss: 7859.4346 - mae: 7859.4346 Epoch 5/100 34/34 [==============================] - 0s 1ms/step - loss: 7639.6699 - mae: 7639.6699 Epoch 6/100 34/34 [==============================] - 0s 2ms/step - loss: 7578.0850 - mae: 7578.0850 Epoch 7/100 34/34 [==============================] - 0s 2ms/step - loss: 7514.6177 - mae: 7514.6177 Epoch 8/100 34/34 [==============================] - 0s 1ms/step - loss: 7694.1338 - mae: 7694.1338 Epoch 9/100 34/34 [==============================] - 0s 1ms/step - loss: 7595.9136 - mae: 7595.9136 Epoch 10/100 34/34 [==============================] - 0s 2ms/step - loss: 7735.9116 - mae: 7735.9116 Epoch 11/100 34/34 [==============================] - 0s 2ms/step - loss: 7444.4189 - mae: 7444.4189 Epoch 12/100 34/34 [==============================] - 0s 2ms/step - loss: 7678.0337 - mae: 7678.0337 Epoch 13/100 34/34 [==============================] - 0s 2ms/step - loss: 7681.5840 - mae: 7681.5840 Epoch 14/100 34/34 [==============================] - 0s 1ms/step - loss: 7702.2842 - mae: 7702.2842 Epoch 15/100 34/34 [==============================] - 0s 1ms/step - loss: 7585.8921 - mae: 7585.8921 Epoch 16/100 34/34 [==============================] - 0s 1ms/step - loss: 7689.5356 - mae: 7689.5356 Epoch 17/100 34/34 [==============================] - 0s 1ms/step - loss: 7509.2036 - mae: 7509.2036 Epoch 18/100 34/34 [==============================] - 0s 1ms/step - loss: 7695.0083 - mae: 7695.0083 Epoch 19/100 34/34 [==============================] - 0s 2ms/step - loss: 7669.3740 - mae: 7669.3740 Epoch 20/100 34/34 [==============================] - 0s 1ms/step - loss: 7901.1362 - mae: 7901.1362 Epoch 21/100 34/34 [==============================] - 0s 2ms/step - loss: 7552.4814 - mae: 7552.4814 Epoch 22/100 34/34 [==============================] - 0s 1ms/step - loss: 7844.9961 - mae: 7844.9961 Epoch 23/100 34/34 [==============================] - 0s 2ms/step - loss: 7642.2485 - mae: 7642.2485 Epoch 24/100 34/34 [==============================] - 0s 2ms/step - loss: 7515.3081 - mae: 7515.3081 Epoch 25/100 34/34 [==============================] - 0s 2ms/step - loss: 7678.3506 - mae: 7678.3506 Epoch 26/100 34/34 [==============================] - 0s 2ms/step - loss: 7653.0269 - mae: 7653.0269 Epoch 27/100 34/34 [==============================] - 0s 2ms/step - loss: 7559.5449 - mae: 7559.5449 Epoch 28/100 34/34 [==============================] - 0s 2ms/step - loss: 7459.9404 - mae: 7459.9404 Epoch 29/100 34/34 [==============================] - 0s 2ms/step - loss: 7618.6177 - mae: 7618.6177 Epoch 30/100 34/34 [==============================] - 0s 1ms/step - loss: 7628.6255 - mae: 7628.6255 Epoch 31/100 34/34 [==============================] - 0s 1ms/step - loss: 7540.4893 - mae: 7540.4893 Epoch 32/100 34/34 [==============================] - 0s 1ms/step - loss: 7486.0186 - mae: 7486.0186 Epoch 33/100 34/34 [==============================] - 0s 2ms/step - loss: 7418.6646 - mae: 7418.6646 Epoch 34/100 34/34 [==============================] - 0s 2ms/step - loss: 7480.7319 - mae: 7480.7319 Epoch 35/100 34/34 [==============================] - 0s 2ms/step - loss: 7615.3115 - mae: 7615.3115 Epoch 36/100 34/34 [==============================] - 0s 1ms/step - loss: 7566.7896 - mae: 7566.7896 Epoch 37/100 34/34 [==============================] - 0s 2ms/step - loss: 7661.0879 - mae: 7661.0879 Epoch 38/100 34/34 [==============================] - 0s 2ms/step - loss: 7522.6816 - mae: 7522.6816 Epoch 39/100 34/34 [==============================] - 0s 2ms/step - loss: 7556.0718 - mae: 7556.0718 Epoch 40/100 34/34 [==============================] - 0s 2ms/step - loss: 7433.5669 - mae: 7433.5669 Epoch 41/100 34/34 [==============================] - 0s 1ms/step - loss: 7722.4312 - mae: 7722.4312 Epoch 42/100 34/34 [==============================] - 0s 2ms/step - loss: 7344.2700 - mae: 7344.2700 Epoch 43/100 34/34 [==============================] - 0s 2ms/step - loss: 7597.4331 - mae: 7597.4331 Epoch 44/100 34/34 [==============================] - 0s 2ms/step - loss: 7338.0132 - mae: 7338.0132 Epoch 45/100 34/34 [==============================] - 0s 2ms/step - loss: 7510.3467 - mae: 7510.3467 Epoch 46/100 34/34 [==============================] - 0s 2ms/step - loss: 7413.5801 - mae: 7413.5801 Epoch 47/100 34/34 [==============================] - 0s 2ms/step - loss: 7451.0391 - mae: 7451.0391 Epoch 48/100 34/34 [==============================] - 0s 2ms/step - loss: 7340.5381 - mae: 7340.5381 Epoch 49/100 34/34 [==============================] - 0s 2ms/step - loss: 7481.9976 - mae: 7481.9976 Epoch 50/100 34/34 [==============================] - 0s 1ms/step - loss: 7468.2842 - mae: 7468.2842 Epoch 51/100 34/34 [==============================] - 0s 1ms/step - loss: 7411.3408 - mae: 7411.3408 Epoch 52/100 34/34 [==============================] - 0s 1ms/step - loss: 7460.0796 - mae: 7460.0796 Epoch 53/100 34/34 [==============================] - 0s 1ms/step - loss: 7601.6606 - mae: 7601.6606 Epoch 54/100 34/34 [==============================] - 0s 2ms/step - loss: 7241.2549 - mae: 7241.2549 Epoch 55/100 34/34 [==============================] - 0s 1ms/step - loss: 7539.6953 - mae: 7539.6953 Epoch 56/100 34/34 [==============================] - 0s 2ms/step - loss: 7293.2012 - mae: 7293.2012 Epoch 57/100 34/34 [==============================] - 0s 2ms/step - loss: 7417.9731 - mae: 7417.9731 Epoch 58/100 34/34 [==============================] - 0s 1ms/step - loss: 7353.0625 - mae: 7353.0625 Epoch 59/100 34/34 [==============================] - 0s 1ms/step - loss: 7643.8247 - mae: 7643.8247 Epoch 60/100 34/34 [==============================] - 0s 2ms/step - loss: 7410.4004 - mae: 7410.4004 Epoch 61/100 34/34 [==============================] - 0s 2ms/step - loss: 7612.8330 - mae: 7612.8330 Epoch 62/100 34/34 [==============================] - 0s 2ms/step - loss: 7387.9087 - mae: 7387.9087 Epoch 63/100 34/34 [==============================] - 0s 2ms/step - loss: 7359.5605 - mae: 7359.5605 Epoch 64/100 34/34 [==============================] - 0s 2ms/step - loss: 7109.0884 - mae: 7109.0884 Epoch 65/100 34/34 [==============================] - 0s 2ms/step - loss: 7396.3223 - mae: 7396.3223 Epoch 66/100 34/34 [==============================] - 0s 2ms/step - loss: 7179.8613 - mae: 7179.8613 Epoch 67/100 34/34 [==============================] - 0s 1ms/step - loss: 7289.7710 - mae: 7289.7710 Epoch 68/100 34/34 [==============================] - 0s 1ms/step - loss: 7523.6973 - mae: 7523.6973 Epoch 69/100 34/34 [==============================] - 0s 1ms/step - loss: 7442.6157 - mae: 7442.6157 Epoch 70/100 34/34 [==============================] - 0s 1ms/step - loss: 7673.4834 - mae: 7673.4834 Epoch 71/100 34/34 [==============================] - 0s 2ms/step - loss: 7276.0337 - mae: 7276.0337 Epoch 72/100 34/34 [==============================] - 0s 2ms/step - loss: 7246.3721 - mae: 7246.3721 Epoch 73/100 34/34 [==============================] - 0s 1ms/step - loss: 7372.0713 - mae: 7372.0713 Epoch 74/100 34/34 [==============================] - 0s 1ms/step - loss: 7512.0762 - mae: 7512.0762 Epoch 75/100 34/34 [==============================] - 0s 2ms/step - loss: 7269.7437 - mae: 7269.7437 Epoch 76/100 34/34 [==============================] - 0s 2ms/step - loss: 7199.5039 - mae: 7199.5039 Epoch 77/100 34/34 [==============================] - 0s 2ms/step - loss: 7261.2920 - mae: 7261.2920 Epoch 78/100 34/34 [==============================] - 0s 2ms/step - loss: 7185.7627 - mae: 7185.7627 Epoch 79/100 34/34 [==============================] - 0s 1ms/step - loss: 7301.7495 - mae: 7301.7495 Epoch 80/100 34/34 [==============================] - 0s 2ms/step - loss: 7002.6309 - mae: 7002.6309 Epoch 81/100 34/34 [==============================] - 0s 1ms/step - loss: 7289.1357 - mae: 7289.1357 Epoch 82/100 34/34 [==============================] - 0s 1ms/step - loss: 7155.3945 - mae: 7155.3945 Epoch 83/100 34/34 [==============================] - 0s 1ms/step - loss: 7475.1709 - mae: 7475.1709 Epoch 84/100 34/34 [==============================] - 0s 1ms/step - loss: 7387.3672 - mae: 7387.3672 Epoch 85/100 34/34 [==============================] - 0s 2ms/step - loss: 7289.9458 - mae: 7289.9458 Epoch 86/100 34/34 [==============================] - 0s 1ms/step - loss: 7268.0942 - mae: 7268.0942 Epoch 87/100 34/34 [==============================] - 0s 2ms/step - loss: 7238.5869 - mae: 7238.5869 Epoch 88/100 34/34 [==============================] - 0s 2ms/step - loss: 7201.7354 - mae: 7201.7349 Epoch 89/100 34/34 [==============================] - 0s 2ms/step - loss: 7538.0757 - mae: 7538.0757 Epoch 90/100 34/34 [==============================] - 0s 2ms/step - loss: 6967.1187 - mae: 6967.1187 Epoch 91/100 34/34 [==============================] - 0s 2ms/step - loss: 7314.1299 - mae: 7314.1299 Epoch 92/100 34/34 [==============================] - 0s 1ms/step - loss: 7192.3115 - mae: 7192.3115 Epoch 93/100 34/34 [==============================] - 0s 1ms/step - loss: 7530.8770 - mae: 7530.8770 Epoch 94/100 34/34 [==============================] - 0s 1ms/step - loss: 7187.3579 - mae: 7187.3579 Epoch 95/100 34/34 [==============================] - 0s 1ms/step - loss: 7561.5635 - mae: 7561.5635 Epoch 96/100 34/34 [==============================] - 0s 1ms/step - loss: 7263.4648 - mae: 7263.4648 Epoch 97/100 34/34 [==============================] - 0s 1ms/step - loss: 7146.2896 - mae: 7146.2896 Epoch 98/100 34/34 [==============================] - 0s 2ms/step - loss: 7247.9238 - mae: 7247.9238 Epoch 99/100 34/34 [==============================] - 0s 2ms/step - loss: 7200.6694 - mae: 7200.6694 Epoch 100/100 34/34 [==============================] - 0s 1ms/step - loss: 7301.6870 - mae: 7301.6870
Out[92]:
<keras.callbacks.History at 0x7f8dedf0cad0>
In [93]:
# Check the results of the insurance model insurance_model.evaluate(X_test, y_test)
# Check the results of the insurance model
insurance_model.evaluate(X_test, y_test)
9/9 [==============================] - 0s 2ms/step - loss: 8628.2393 - mae: 8628.2393
Out[93]:
[8628.2392578125, 8628.2392578125]
Our model didn’t perform very well, let’s try a bigger model.
We’ll try 3 things:
- Increasing the number of layers (2 -> 3).
- Increasing the number of units in each layer (except for the output layer).
- Changing the optimizer (from SGD to Adam).
Everything else will stay the same.
In [94]:
# Set random seed tf.random.set_seed(42) # Add an extra layer and increase number of units insurance_model_2 = tf.keras.Sequential([ tf.keras.layers.Dense(100), # 100 units tf.keras.layers.Dense(10), # 10 units tf.keras.layers.Dense(1) # 1 unit (important for output layer) ]) # Compile the model insurance_model_2.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.Adam(), # Adam works but SGD doesn't metrics=['mae']) # Fit the model and save the history (we can plot this) history = insurance_model_2.fit(X_train, y_train, epochs=100, verbose=0)
# Set random seed
tf.random.set_seed(42)
# Add an extra layer and increase number of units
insurance_model_2 = tf.keras.Sequential([
tf.keras.layers.Dense(100), # 100 units
tf.keras.layers.Dense(10), # 10 units
tf.keras.layers.Dense(1) # 1 unit (important for output layer)
])
# Compile the model
insurance_model_2.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.Adam(), # Adam works but SGD doesn’t
metrics=[‘mae’])
# Fit the model and save the history (we can plot this)
history = insurance_model_2.fit(X_train, y_train, epochs=100, verbose=0)
In [95]:
# Evaluate our larger model insurance_model_2.evaluate(X_test, y_test)
# Evaluate our larger model
insurance_model_2.evaluate(X_test, y_test)
9/9 [==============================] - 0s 2ms/step - loss: 4924.3477 - mae: 4924.3477
Out[95]:
[4924.34765625, 4924.34765625]
Much better! Using a larger model and the Adam optimizer results in almost half the error as the previous model.
🔑 Note: For many problems, the Adam optimizer is a great starting choice. See Andrei Karpathy’s «Adam is safe» point from A Recipe for Training Neural Networks for more.
Let’s check out the loss curves of our model, we should see a downward trend.
In [96]:
# Plot history (also known as a loss curve) pd.DataFrame(history.history).plot() plt.ylabel("loss") plt.xlabel("epochs");
# Plot history (also known as a loss curve)
pd.DataFrame(history.history).plot()
plt.ylabel(«loss»)
plt.xlabel(«epochs»);
From this, it looks like our model’s loss (and MAE) were both still decreasing (in our case, MAE and loss are the same, hence the lines in the plot overlap eachother).
What this tells us is the loss might go down if we try training it for longer.
🤔 Question: How long should you train for?
It depends on what problem you’re working on. Sometimes training won’t take very long, other times it’ll take longer than you expect. A common method is to set your model training for a very long time (e.g. 1000’s of epochs) but set it up with an EarlyStopping callback so it stops automatically when it stops improving. We’ll see this in another module.
Let’s train the same model as above for a little longer. We can do this but calling fit on it again.
In [97]:
# Try training for a little longer (100 more epochs) history_2 = insurance_model_2.fit(X_train, y_train, epochs=100, verbose=0)
# Try training for a little longer (100 more epochs)
history_2 = insurance_model_2.fit(X_train, y_train, epochs=100, verbose=0)
How did the extra training go?
In [98]:
# Evaluate the model trained for 200 total epochs insurance_model_2_loss, insurance_model_2_mae = insurance_model_2.evaluate(X_test, y_test) insurance_model_2_loss, insurance_model_2_mae
# Evaluate the model trained for 200 total epochs
insurance_model_2_loss, insurance_model_2_mae = insurance_model_2.evaluate(X_test, y_test)
insurance_model_2_loss, insurance_model_2_mae
9/9 [==============================] - 0s 2ms/step - loss: 3494.7285 - mae: 3494.7285
Out[98]:
(3494.728515625, 3494.728515625)
Boom! Training for an extra 100 epochs we see about a 10% decrease in error.
How does the visual look?
In [99]:
# Plot the model trained for 200 total epochs loss curves pd.DataFrame(history_2.history).plot() plt.ylabel("loss") plt.xlabel("epochs"); # note: epochs will only show 100 since we overrid the history variable
# Plot the model trained for 200 total epochs loss curves
pd.DataFrame(history_2.history).plot()
plt.ylabel(«loss»)
plt.xlabel(«epochs»); # note: epochs will only show 100 since we overrid the history variable
Preprocessing data (normalization and standardization)¶
A common practice when working with neural networks is to make sure all of the data you pass to them is in the range 0 to 1.
This practice is called normalization (scaling all values from their original range to, e.g. between 0 and 100,000 to be between 0 and 1).
There is another process call standardization which converts all of your data to unit variance and 0 mean.
These two practices are often part of a preprocessing pipeline (a series of functions to prepare your data for use with neural networks).
Knowing this, some of the major steps you’ll take to preprocess your data for a neural network include:
- Turning all of your data to numbers (a neural network can’t handle strings).
- Making sure your data is in the right shape (verifying input and output shapes).
- Feature scaling:
- Normalizing data (making sure all values are between 0 and 1). This is done by subtracting the minimum value then dividing by the maximum value minus the minmum. This is also referred to as min-max scaling.
- Standardization (making sure all values have a mean of 0 and a variance of 1). This is done by substracting the mean value from the target feature and then dividing it by the standard deviation.
- Which one should you use?
- With neural networks you’ll tend to favour normalization as they tend to prefer values between 0 and 1 (you’ll see this espcially with image processing), however, you’ll often find a neural network can perform pretty well with minimal feature scaling.
📖 Resource: For more on preprocessing data, I’d recommend reading the following resources:
- Scikit-Learn’s documentation on preprocessing data.
- Scale, Standardize or Normalize with Scikit-Learn by Jeff Hale.
We’ve already turned our data into numbers using get_dummies(), let’s see how we’d normalize it as well.
In [100]:
import pandas as pd import matplotlib.pyplot as plt import tensorflow as tf # Read in the insurance dataset insurance = pd.read_csv("https://raw.githubusercontent.com/stedy/Machine-Learning-with-R-datasets/master/insurance.csv")
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
# Read in the insurance dataset
insurance = pd.read_csv(«https://raw.githubusercontent.com/stedy/Machine-Learning-with-R-datasets/master/insurance.csv»)
In [101]:
# Check out the data insurance.head()
# Check out the data
insurance.head()
Out[101]:
| age | sex | bmi | children | smoker | region | charges | |
|---|---|---|---|---|---|---|---|
| 0 | 19 | female | 27.900 | 0 | yes | southwest | 16884.92400 |
| 1 | 18 | male | 33.770 | 1 | no | southeast | 1725.55230 |
| 2 | 28 | male | 33.000 | 3 | no | southeast | 4449.46200 |
| 3 | 33 | male | 22.705 | 0 | no | northwest | 21984.47061 |
| 4 | 32 | male | 28.880 | 0 | no | northwest | 3866.85520 |
Now, just as before, we need to transform the non-numerical columns into numbers and this time we’ll also be normalizing the numerical columns with different ranges (to make sure they’re all between 0 and 1).
To do this, we’re going to use a few classes from Scikit-Learn:
make_column_transformer— build a multi-step data preprocessing function for the folllowing transformations:MinMaxScaler— make sure all numerical columns are normalized (between 0 and 1).OneHotEncoder— one hot encode the non-numerical columns.
Let’s see them in action.
In [102]:
from sklearn.compose import make_column_transformer from sklearn.preprocessing import MinMaxScaler, OneHotEncoder # Create column transformer (this will help us normalize/preprocess our data) ct = make_column_transformer( (MinMaxScaler(), ["age", "bmi", "children"]), # get all values between 0 and 1 (OneHotEncoder(handle_unknown="ignore"), ["sex", "smoker", "region"]) ) # Create X & y X = insurance.drop("charges", axis=1) y = insurance["charges"] # Build our train and test sets (use random state to ensure same split as before) X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42) # Fit column transformer on the training data only (doing so on test data would result in data leakage) ct.fit(X_train) # Transform training and test data with normalization (MinMaxScalar) and one hot encoding (OneHotEncoder) X_train_normal = ct.transform(X_train) X_test_normal = ct.transform(X_test)
from sklearn.compose import make_column_transformer
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder
# Create column transformer (this will help us normalize/preprocess our data)
ct = make_column_transformer(
(MinMaxScaler(), [«age», «bmi», «children»]), # get all values between 0 and 1
(OneHotEncoder(handle_unknown=»ignore»), [«sex», «smoker», «region»])
)
# Create X & y
X = insurance.drop(«charges», axis=1)
y = insurance[«charges»]
# Build our train and test sets (use random state to ensure same split as before)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Fit column transformer on the training data only (doing so on test data would result in data leakage)
ct.fit(X_train)
# Transform training and test data with normalization (MinMaxScalar) and one hot encoding (OneHotEncoder)
X_train_normal = ct.transform(X_train)
X_test_normal = ct.transform(X_test)
Now we’ve normalized it and one-hot encoding it, what does our data look like now?
In [103]:
# Non-normalized and non-one-hot encoded data example X_train.loc[0]
# Non-normalized and non-one-hot encoded data example
X_train.loc[0]
Out[103]:
age 19 sex female bmi 27.9 children 0 smoker yes region southwest Name: 0, dtype: object
In [104]:
# Normalized and one-hot encoded example X_train_normal[0]
# Normalized and one-hot encoded example
X_train_normal[0]
Out[104]:
array([0.60869565, 0.10734463, 0.4 , 1. , 0. ,
1. , 0. , 0. , 1. , 0. ,
0. ])
In [105]:
# Notice the normalized/one-hot encoded shape is larger because of the extra columns X_train_normal.shape, X_train.shape
# Notice the normalized/one-hot encoded shape is larger because of the extra columns
X_train_normal.shape, X_train.shape
Our data is normalized and numerical, let’s model it.
We’ll use the same model as insurance_model_2.
In [106]:
# Set random seed tf.random.set_seed(42) # Build the model (3 layers, 100, 10, 1 units) insurance_model_3 = tf.keras.Sequential([ tf.keras.layers.Dense(100), tf.keras.layers.Dense(10), tf.keras.layers.Dense(1) ]) # Compile the model insurance_model_3.compile(loss=tf.keras.losses.mae, optimizer=tf.keras.optimizers.Adam(), metrics=['mae']) # Fit the model for 200 epochs (same as insurance_model_2) insurance_model_3.fit(X_train_normal, y_train, epochs=200, verbose=0)
# Set random seed
tf.random.set_seed(42)
# Build the model (3 layers, 100, 10, 1 units)
insurance_model_3 = tf.keras.Sequential([
tf.keras.layers.Dense(100),
tf.keras.layers.Dense(10),
tf.keras.layers.Dense(1)
])
# Compile the model
insurance_model_3.compile(loss=tf.keras.losses.mae,
optimizer=tf.keras.optimizers.Adam(),
metrics=[‘mae’])
# Fit the model for 200 epochs (same as insurance_model_2)
insurance_model_3.fit(X_train_normal, y_train, epochs=200, verbose=0)
Out[106]:
<keras.callbacks.History at 0x7f8df4a6da50>
Let’s evaluate the model on normalized test set.
In [107]:
# Evaulate 3rd model insurance_model_3_loss, insurance_model_3_mae = insurance_model_3.evaluate(X_test_normal, y_test)
# Evaulate 3rd model
insurance_model_3_loss, insurance_model_3_mae = insurance_model_3.evaluate(X_test_normal, y_test)
9/9 [==============================] - 0s 2ms/step - loss: 3171.5774 - mae: 3171.5774
And finally, let’s compare the results from insurance_model_2 (trained on non-normalized data) and insurance_model_3 (trained on normalized data).
In [108]:
# Compare modelling results from non-normalized data and normalized data insurance_model_2_mae, insurance_model_3_mae
# Compare modelling results from non-normalized data and normalized data
insurance_model_2_mae, insurance_model_3_mae
Out[108]:
(3494.728515625, 3171.577392578125)
From this we can see normalizing the data results in 10% less error using the same model than not normalizing the data.
This is one of the main benefits of normalization: faster convergence time (a fancy way of saying, your model gets to better results faster).
insurance_model_2 may have eventually achieved the same results as insurance_model_3 if we left it training for longer.
Also, the results may change if we were to alter the architectures of the models, e.g. more hidden units per layer or more layers.
But since our main goal as neural network practitioners is to decrease the time between experiments, anything that helps us get better results sooner is a plus.
🛠 Exercises¶
We’ve a covered a whole lot pretty quickly.
So now it’s time to have a play around with a few things and start to build up your intuition.
I emphasise the words play around because that’s very important. Try a few things out, run the code and see what happens.
- Create your own regression dataset (or make the one we created in «Create data to view and fit» bigger) and build fit a model to it.
- Try building a neural network with 4 Dense layers and fitting it to your own regression dataset, how does it perform?
- Try and improve the results we got on the insurance dataset, some things you might want to try include:
- Building a larger model (how does one with 4 dense layers go?).
- Increasing the number of units in each layer.
- Lookup the documentation of Adam and find out what the first parameter is, what happens if you increase it by 10x?
- What happens if you train for longer (say 300 epochs instead of 200)?
- Import the Boston pricing dataset from TensorFlow
tf.keras.datasetsand model it.