Содержание
- Python Pandas ошибка токенизации данных
- 19 ответов:
- Python read_csv — ParserError: Error tokenizing data
- 2 Answers 2
- Python 3 Pandas Error: pandas.parser.CParserError: Error tokenizing data. C error: Expected 11 fields in line 5, saw 13
- 8 Answers 8
- Linked
- Related
- Hot Network Questions
- Subscribe to RSS
- read_csv «CParserError: Error tokenizing data» with variable number of fields #11493
- Comments
Python Pandas ошибка токенизации данных
Я пытаюсь использовать панды для манипулирования .CSV-файл, но я получаю эту ошибку:
панды.синтаксический анализатор.CParserError: ошибка маркирования данных. C ошибка: ожидается 2 поля в строке 3, увидел 12
Я попытался прочитать документы панды, но ничего не нашел.
мой код очень простой:
Как я могу решить это? Я должен использовать csv модуль или другой язык ?
19 ответов:
- разделители в
- первая строка, как отметил @TomAugspurger
чтобы решить эту проблему, попробуйте указать sep и/или header аргументов при вызове read_csv . Например,
в коде выше, sep определяет ваш разделитель и header=None говорит панд, что ваши исходные данные не имеют строки для заголовков / заголовков столбцов. Так говорит документы: «если файл не содержит строки заголовка, то вы должны явно передать header=None». В этом случае pandas автоматически создает целочисленные индексы для каждого поля <0,1,2. >.
согласно документам, разделитель вещь должна не быть проблемой. В документах говорится, что » если sep нет [не указано], попытается автоматически определить это.»Мне, однако, не повезло с этим, включая случаи с очевидными разделителями.
синтаксический анализатор запутывается в заголовке файла. Он читает первую строку и выводит количество столбцов из этой строки. Но первые две строки не являются репрезентативными для фактических данных в файле.
попробуйте data = pd.read_csv(path, skiprows=2)
CSV-файл может иметь переменное количество столбцов и read_csv выведено количество столбцов из первых нескольких строк. Два способа решить ее в этом случае:
1) измените файл CSV на фиктивную первую строку с максимальным количеством столбцов (и укажите header=[0] )
2) или использовать names = list(range(0,N)) где N-максимальное количество столбцов.
у меня тоже была эта проблема, но, возможно, по другой причине. У меня были некоторые конечные запятые в моем CSV, которые добавляли дополнительный столбец, который панды пытались прочитать. Используя следующие работы, но он просто игнорирует плохие строки:
Если вы хотите сохранить линии уродливый вид рубить для обработки ошибок, чтобы сделать что-то вроде следующего:
я приступил к написанию сценария для повторной вставки строк в фрейм данных, так как плохие строки будут заданы переменной ‘line’ в приведенном выше коде. Этого можно избежать, просто используя читатель csv. Будем надеяться, что разработчики панды могут сделать его легче справиться с этой ситуацией в будущем.
это, безусловно, вопрос разделителя, так как большинство csv CSV получают создать с помощью sep=’/t’ так попробуй read_csv используя символ табуляции (t) С помощью разделителя /t . Итак, попробуйте открыть с помощью следующей строки кода.
У меня была эта проблема несколько раз. Почти каждый раз, причина в том, что файл, который я пытался открыть, не был правильно сохранен CSV для начала. И под «правильно» я имею в виду, что каждая строка имела одинаковое количество разделителей или столбцов.
обычно это происходило потому, что я открыл CSV в Excel, а затем неправильно сохранил его. Несмотря на то, что расширение файла все еще было .csv, чистый формат CSV был изменен.
любой файл, сохраненный с pandas to_csv будет правильно отформатирован и не должен иметь этой проблемы. Но если вы откроете его с другой программой, это может изменить структуру.
надеюсь, что это поможет.
я столкнулся с той же проблемой. Используя pd.read_table() на том же исходном файле, казалось, работали. Я не мог проследить причину этого, но это было полезным обходным путем для моего случая. Возможно, кто-то более знающий может пролить свет на то, почему это сработало.
изменить: Я обнаружил, что эта ошибка ползет вверх, когда у вас есть какой-то текст в файле, который не имеет того же формата, что и фактические данные. Обычно это информация верхнего или нижнего колонтитула (больше одной строки, поэтому skip_header не работает) который не будет разделен тем же количеством запятых, что и ваши фактические данные (при использовании read_csv). Использование read_table использует вкладку в качестве разделителя, который может обойти текущую ошибку пользователей, но ввести другие.
обычно я обхожу это, читая дополнительные данные в файл, а затем использую метод read_csv ().
точное решение может отличаться в зависимости от вашего фактического файла, но этот подход работал для меня в нескольких случаях
у меня была аналогичная проблема при попытке прочитать таблицу с разделителями табуляции с пробелами, запятыми и кавычками:
Это говорит, что это имеет какое-то отношение к C parsing engine (который является стандартным). Может быть, переход на python один изменит что-нибудь
теперь это другая ошибка.
Если мы продолжим и попытаемся удалить пробелы из таблицы, ошибка из python-engine снова изменится:
и это становится ясно что у панд были проблемы с разбором наших строк. Чтобы разобрать таблицу с движком python, мне нужно было удалить все пробелы и кавычки из таблицы заранее. Тем временем C-engine продолжал рушиться даже с запятыми в строках.
Чтобы избежать создания нового файла с заменами я сделал это, так как мои таблицы малы:
tl; dr
Измените механизм синтаксического анализа, старайтесь избегать любых не разделяющих кавычек / запятых / пробелов в ваших данных.
хотя это не относится к этому вопросу, эта ошибка также может появиться со сжатыми данными. Явно устанавливая значение для kwarg compression решена моя проблема.
Иногда проблема заключается не в том, как использовать python, а с необработанными данными.
Я получил это сообщение об ошибке
оказалось, что в описании столбца иногда встречаются запятые. Это означает, что CSV-файл необходимо очистить или использовать другой разделитель.
при попытке прочитать данные csv из ссылки
Я скопировал данные с сайта в свой csvfile. У него были дополнительные пробелы, поэтому он использовал sep =’, ‘ и он работал 🙂
альтернатива, которую я нашел полезной при работе с подобными ошибками синтаксического анализа, использует модуль CSV для перенаправления данных в pandas df. Например:
Я считаю, что модуль CSV немного более надежен для плохо отформатированных файлов, разделенных запятыми, и поэтому имел успех с этим маршрутом для решения таких проблем.
У меня был набор данных с предварительными номерами строк, я использовал index_col:
следующая последовательность команд работает (я теряю первую строку данных-нет заголовка=нет настоящего -, но по крайней мере он загружается):
df = pd.read_csv(filename, usecols=range(0, 42)) df.columns = [‘YR’, ‘MO’, ‘DAY’, ‘HR’, ‘MIN’, ‘SEC’, ‘HUND’, ‘ERROR’, ‘RECTYPE’, ‘LANE’, ‘SPEED’, ‘CLASS’, ‘LENGTH’, ‘GVW’, ‘ESAL’, ‘W1’, ‘S1’, ‘W2’, ‘S2’, ‘W3’, ‘S3’, ‘W4’, ‘S4’, ‘W5’, ‘S5’, ‘W6’, ‘S6’, ‘W7’, ‘S7’, ‘W8’, ‘S8’, ‘W9’, ‘S9’, ‘W10’, ‘S10’, ‘W11’, ‘S11’, ‘W12’, ‘S12’, ‘W13’, ‘S13’, ‘W14’]
следующее не работает:
df = pd.read_csv(filename, names=[‘YR’, ‘MO’, ‘DAY’, ‘HR’, ‘MIN’, ‘SEC’, ‘HUND’, ‘ERROR’, ‘RECTYPE’, ‘LANE’, ‘SPEED’, ‘CLASS’, ‘LENGTH’, ‘GVW’, ‘ESAL’, ‘W1’, ‘S1’, ‘W2’, ‘S2’, ‘W3’, ‘S3’, ‘W4’, ‘S4’, ‘W5’, ‘S5’, ‘W6’, ‘S6’, ‘W7’, ‘S7’, ‘W8’, ‘S8’, ‘W9’, ‘S9’, ‘W10’, ‘S10’, ‘W11’, ‘S11’, ‘W12’, ‘S12’, ‘W13’, ‘S13’, ‘W14’], usecols=range(0, 42))
CParserError: ошибка маркирования данных. C ошибка: ожидается 53 поля в строке 1605634, увидел 54 Следующее не работает:
df = pd.read_csv(filename, header=None)
CParserError: ошибка маркирования данных. C ошибка: ожидается 53 поля в строке 1605634, saw 54
следовательно, в вашей проблеме вы должны пройти usecols=range(0, 2)
У меня была аналогичная ошибка, и проблема заключалась в том, что у меня были некоторые экранированные кавычки в моем csv-файле и нужно было установить параметр escapechar соответствующим образом.
Вы можете сделать этот шаг, чтобы избежать проблемы —
Источник
Python read_csv — ParserError: Error tokenizing data
I understand why I get this error when trying to df = pd.read_csv(file) :
ParserError: Error tokenizing data. C error: Expected 14 fields in line 7, saw 30
When it reads in the csv, it sees 14 strings/columns in the first row, based on the first row of the csv calls it the headers (which is what I want).
However, those columns are extended further, down the rows (specifially when it gets to row 7).
I can find solutions that will read it in by skipping those rows 1-6, but I don’t want that. I still want the whole csv to be read, but instead of the header being 14 columns, how can I tell it make the header 30 columns, and if there is no text/string, just leave the column as a «», or null, or some random numbering. In other words, I don’t care what it’s named, I just need the space holder so it can parse after row 6.
I’m wondering is there a way to read in the csv, and explicitly say there are 30 columns but have not found a solution.
2 Answers 2
I can throw some random solutions that I think should work.
1) Set Header=None and give columns names in ‘Name’ attribute of read_csv.
PS. This will work if your CSV doesn’t have a header already.
2) Secondly you can try using below command (if your csv already has header row)
Let me know if this works out for you.
what about trying, to be noted error_bad_lines=False will cause the offending lines to be skipped
Just few more collectives answers..
It might be an issue with the delimiters in your data the first row, To solve it, try specifying the sep and/or header arguments when calling read_csv. For instance,
In the code above, sep defines your delimiter and header=None tells pandas that your source data has no row for headers / column titles. Here Documenet says: «If file contains no header row, then you should explicitly pass header=None». In this instance, pandas automatically creates whole-number indices for each field <0,1,2. >.
According to the docs, the delimiter thing should not be an issue. The docs say that «if sep is None [not specified], will try to automatically determine this.» I however have not had good luck with this, including instances with obvious delimiters.
This might be an issue of delimiter, as most of the csv CSV are got create using sep=’/t’ so try to read_csv using the tab character (t) using separator /t. so, try to open using following code line.
Источник
Python 3 Pandas Error: pandas.parser.CParserError: Error tokenizing data. C error: Expected 11 fields in line 5, saw 13
I checked out this answer as I am having a similar problem.
However, for some reason ALL of my rows are being skipped.
My code is simple:
and the error I get is:
8 Answers 8
Solution is to use pandas built-in delimiter «sniffing».
For those landing here, I got this error when the file was actually an .xls file not a true .csv. Try resaving as a csv in a spreadsheet app.
I had the same error, I read my csv data using this : d1 = pd.read_json(‘my.csv’) then I try this d1 = pd.read_json(‘my.csv’, sep=’t’) and this time it’s right. So you could try this method if your delimiter is not ‘,’, because the default is ‘,’, so if you don’t indicate clearly, it go wrong. pandas.read_csv
This error means, you get unequal number of columns for each row. In your case, until row 5, you’ve had 11 columns but in line 5 you have 13 inputs (columns).
For this problem, you can try the following approach to open read your file:
This parsing error could occur for multiple reasons and solutions to the different reasons have been posted here as well as in Python Pandas Error tokenizing data.
I posted a solution to one possible reason for this error here: https://stackoverflow.com/a/43145539/6466550
I have had similar problems. With my csv files it occurs because they were created in R , so it has some extra commas and different spacing than a «regular» csv file.
I found that if I did a read.table in R, I could then save it using write.csv and the option of row.names = F .
I could not get any of the read options in pandas to help me.
The problem could be that one or multiple rows of csv file contain more delimiters (commas , ) than expected. It is solved when each row matches the amount of delimiters of the first line of the csv file where the column names are defined.
use t+ in the separator pattern instead of t.
Linked
Hot Network Questions
To subscribe to this RSS feed, copy and paste this URL into your RSS reader.
Site design / logo © 2023 Stack Exchange Inc; user contributions licensed under CC BY-SA . rev 2023.1.14.43159
By clicking “Accept all cookies”, you agree Stack Exchange can store cookies on your device and disclose information in accordance with our Cookie Policy.
Источник
read_csv «CParserError: Error tokenizing data» with variable number of fields #11493
I am having trouble with read_csv (Pandas 0.17.0) when trying to read a 380+ MB csv file. The file starts with 54 fields but some lines have 53 fields instead of 54. Running the below code gives me the following error:
If I pass the error_bad_lines=False keyword, problematic lines are displayed similar to the example below:
however I get the following error this time ( also the DataFrame does not get loaded):
If I pass the engine=’python’ keyword, I do not get any errors, but it takes a really long time to parse the data. Please note that 53 and 54 are switched in the error messages depending on if error_bad_lines=False is used or not.
The text was updated successfully, but these errors were encountered:
these errors are all correct. you are constraining what the parser is doing by passing usecols , and names . don’t do this and see if you can parse it.
very hard to diagnose something like this without a sample of the file that reproduces.
also show pd.show_versions()
With the original data file:
pd.read_csv(filename) with no other keywords seems to parse the data with no errors. pd.read_csv(filename, header=None) gives the following error:
Totally agreed that it is very hard to diagnose without the sample data. I tried generating the error with a csv file with a few lines (some has 53 fields, some 54), pd.read_csv fills the gaps with NaNs as expected. I repeated by passing usecols and header=None , still works. It seems the original file has some kind of an issue that raises all the errors.
pd.show_versions() output as follows:
is expected as the number of columns is inferred from the first line. If you pass names if will use this as a determining feature.
So keep trying various options. You are constraining it a bit much actually with names and usecols . You might be better off reading it in, then reindexing to what you need.
If engine=’python’ is used, curiously, it loads the DataFrame without any hiccups though. I used the following snippet to extract the first 3 lines in the file and 3 of the offending lines (got the line numbers from the error message).
As you suggested, I will try to read the file, then modify the DataFrame (rename columns, delete unnecessary ones etc.) or simply use the python engine (long processing time).
Источник
In this tutorial, we’ll see how to solve a common Pandas read_csv() error – Error Tokenizing Data. The full error is something like:
ParserError: Error tokenizing data. C error: Expected 2 fields in line 4, saw 4
The Pandas parser error when reading csv is very common but difficult to investigate and solve for big CSV files.
There could be many different reasons for this error:
- «wrong» data in the file
- different number of columns
- mixed data
- several data files stored as a single file
- nested separators
- wrong parsing parameters for read_csv()
- different separator
- line terminators
- wrong parsing engine
Let’s cover the steps to diagnose and solve this error
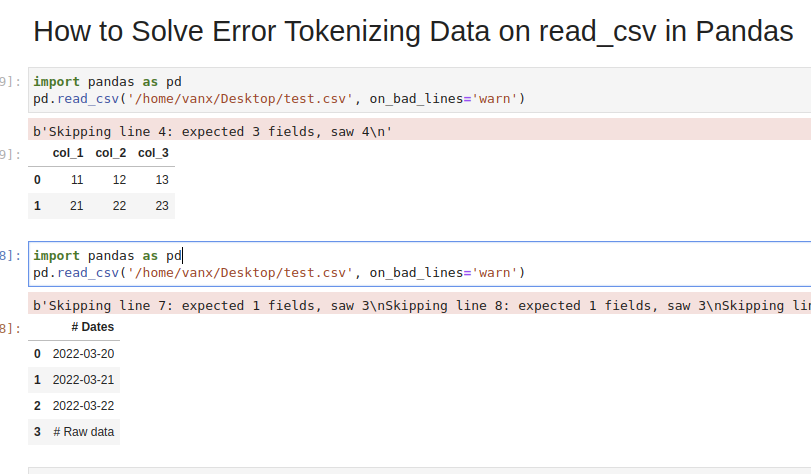
Step 1: Analyze and Skip Bad Lines for Error Tokenizing Data
Suppose we have CSV file like:
col_1,col_2,col_3
11,12,13
21,22,23
31,32,33,44
which we are going to read by — read_csv() method:
import pandas as pd
pd.read_csv('test.csv')
We will get an error:
ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
We can easily see where the problem is. But what should be the solution in this case? Remove the 44 or add a new column? It depends on the context of this data.
If we don’t need the bad data we can use parameter — on_bad_lines='skip' in order to skip bad lines:
pd.read_csv('test.csv', on_bad_lines='skip')
For older Pandas versions you may need to use: error_bad_lines=False which will be deprecated in future.
Using warn instead of skip will produce:
pd.read_csv('test.csv', on_bad_lines='warn')
warning like:
b'Skipping line 4: expected 3 fields, saw 4n'
To find more about how to drop bad lines with read_csv() read the linked article.
Step 2: Use correct separator to solve Pandas tokenizing error
In some cases the reason could be the separator used to read the CSV file. In this case we can open the file and check its content.
If you don’t know how to read huge files in Windows or Linux — then check the article.
Depending on your OS and CSV file you may need to use different parameters like:
sep—lineterminator—engine
More information on the parameters can be found in Pandas doc for read_csv()
If the CSV file has tab as a separator and different line endings we can use:
import pandas as pd
pd.read_csv('test.csv', sep='t', lineterminator='rn')
Note that delimiter is an alias for sep.
Step 3: Use different engine for read_csv()
The default C engine cannot automatically detect the separator, but the python parsing engine can.
There are 3 engines in the latest version of Pandas:
cpythonpyarrow
Python engine is the slowest one but the most feature-complete.
Using python as engine can help detecting the correct delimiter and solve the Error Tokenizing Data:
pd.read_csv('test.csv', engine='python')
Sometimes Error Tokenizing Data problem may be related to the headers.
For example multiline headers or additional data can produce the error. In this case we can skip the first or last few lines by:
pd.read_csv('test.csv', skiprows=1, engine='python')
or skipping the last lines:
pd.read_csv('test.csv', skipfooter=1, engine='python')
Note that we need to use python as the read_csv() engine for parameter — skipfooter.
In some cases header=None can help in order to solve the error:
pd.read_csv('test.csv', header=None)
Step 5: Autodetect skiprows for read_csv()
Suppose we had CSV file like:
Dates
2022-03-20
2022-03-21
2022-03-22
Raw data
col_1,col_2,col_3
11,12,13
21,22,23
31,32,33
We are interested in reading the data stored after the line «Raw data». If this line is changing for different files and we need to autodetect the line.
To search for a line containing some match in Pandas we can use a separator which is not present in the file «@@».
In case of multiple occurrences we can get the biggest index. To autodetect skiprows parameter in Pandas read_csv() use:
df_temp = pd.read_csv('test.csv', sep='@@', engine='python', names=['col'])
ix_last_start = df_temp[df_temp['col'].str.contains('# Raw data')].index.max()
result is:
4
Finding the starting line can be done by visually inspecting the file in the text editor.
Then we can simply read the file by:
df_analytics = pd.read_csv(file, skiprows=ix_last_start)
Conclusion
In this post we saw how to investigate the error:
ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
We covered different reasons and solutions in order to read any CSV file with Pandas.
Some solutions will warn and skip problematic lines. Others will try to resolve the problem automatically.
While reading a CSV file, you may get the “Pandas Error Tokenizing Data“. This mostly occurs due to the incorrect data in the CSV file.
You can solve python pandas error tokenizing data error by ignoring the offending lines using error_bad_lines=False.
In this tutorial, you’ll learn the cause and how to solve the error tokenizing data error.
If you’re in Hurry
You can use the below code snippet to solve the tokenizing error. You can solve the error by ignoring the offending lines and suppressing errors.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', error_bad_lines=False, engine ='python')
dfIf You Want to Understand Details, Read on…
In this tutorial, you’ll learn the causes for the exception “Error Tokenizing Data” and how it can be solved.
Cause of the Problem
- CSV file has two header lines
- Different separator is used
r– is a new line character and it is present in column names which makes subsequent column names to be read as next line- Lines of the CSV files have inconsistent number of columns
In the case of invalid rows which has an inconsistent number of columns, you’ll see an error as Expected 1 field in line 12, saw m. This means it expected only 1 field in the CSV file but it saw 12 values after tokenizing it. Hence, it doesn’t know how the tokenized values need to be handled. You can solve the errors by using one of the options below.
Finding the Problematic Line (Optional)
If you want to identify the line which is creating the problem while reading, you can use the below code snippet.
It uses the CSV reader. hence it is similar to the read_csv() method.
Snippet
import csv
with open("sample.csv", 'rb') as file_obj:
reader = csv.reader(file_obj)
line_no = 1
try:
for row in reader:
line_no += 1
except Exception as e:
print (("Error in the line number %d: %s %s" % (line_no, str(type(e)), e.message)))Using Err_Bad_Lines Parameter
When there is insufficient data in any of the rows, the tokenizing error will occur.
You can skip such invalid rows by using the err_bad_line parameter within the read_csv() method.
This parameter controls what needs to be done when a bad line occurs in the file being read.
If it’s set to,
False– Errors will be suppressed for Invalid linesTrue– Errors will be thrown for invalid lines
Use the below snippet to read the CSV file and ignore the invalid lines. Only a warning will be shown with the line number when there is an invalid lie found.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', error_bad_lines=False)
dfIn this case, the offending lines will be skipped and only the valid lines will be read from CSV and a dataframe will be created.
Using Python Engine
There are two engines supported in reading a CSV file. C engine and Python Engine.
C Engine
- Faster
- Uses C language to parse the CSV file
- Supports
float_precision - Cannot automatically detect the separator
- Doesn’t support skipping footer
Python Engine
- Slower when compared to C engine but its feature complete
- Uses Python language to parse the CSV file
- Doesn’t support
float_precision. Not required with Python - Can automatically detect the separator
- Supports skipping footer
Using the python engine can solve the problems faced while parsing the files.
For example, When you try to parse large CSV files, you may face the Error tokenizing data. c error out of memory. Using the python engine can solve the memory issues while parsing such big CSV files using the read_csv() method.
Use the below snippet to use the Python engine for reading the CSV file.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', engine='python', error_bad_lines=False)
dfThis is how you can use the python engine to parse the CSV file.
Optionally, this could also solve the error Error tokenizing data. c error out of memory when parsing the big CSV files.
Using Proper Separator
CSV files can have different separators such as tab separator or any other special character such as ;. In this case, an error will be thrown when reading the CSV file, if the default C engine is used.
You can parse the file successfully by specifying the separator explicitly using the sep parameter.
As an alternative, you can also use the python engine which will automatically detect the separator and parse the file accordingly.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', sep='t')
dfThis is how you can specify the separator explicitly which can solve the tokenizing errors while reading the CSV files.
Using Line Terminator
CSV file can contain r carriage return for separating the lines instead of the line separator n.
In this case, you’ll face CParserError: Error tokenizing data. C error: Buffer overflow caught - possible malformed input file when the line contains the r instead on n.
You can solve this error by using the line terminator explicitly using the lineterminator parameter.
Snippet
df = pd.read_csv('sample.csv',
lineterminator='n')This is how you can use the line terminator to parse the files with the terminator r.
Using header=None
CSV files can have incomplete headers which can cause tokenizing errors while parsing the file.
You can use header=None to ignore the first line headers while reading the CSV files.
This will parse the CSV file without headers and create a data frame. You can also add headers to column names by adding columns attribute to the read_csv() method.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, error_bad_lines=False)
dfThis is how you can ignore the headers which are incomplete and cause problems while reading the file.
Using Skiprows
CSV files can have headers in more than one row. This can happen when data is grouped into different sections and each group is having a name and has columns in each section.
In this case, you can ignore such rows by using the skiprows parameter. You can pass the number of rows to be skipped and the data will be read after skipping those number of rows.
Use the below snippet to skip the first two rows while reading the CSV file.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, skiprows=2, error_bad_lines=False)
dfThis is how you can skip or ignore the erroneous headers while reading the CSV file.
Reading As Lines and Separating
In a CSV file, you may have a different number of columns in each row. This can occur when some of the columns in the row are considered optional. You may need to parse such files without any problems during tokenizing.
In this case, you can read the file as lines and separate it later using the delimiter and create a dataframe out of it. This is helpful when you have varying lengths of rows.
In the below example,
- the file is read as lines by specifying the separator as a new line using
sep='n'. Now the file will be tokenized on each new line, and a single column will be available in the dataframe. - You can split the lines using the separator or regex and create different columns out of it.
expand=Trueexpands the split string into multiple columns.
Use the below snippet to read the file as lines and separate it using the separator.
Snippet
import pandas as pd
df = pd.read_csv('sample.csv', header=None, sep='n')
df = df[0].str.split('s|s', expand=True)
dfThis is how you can read the file as lines and later separate it to avoid problems while parsing the lines with an inconsistent number of columns.
Conclusion
To summarize, you’ve learned the causes of the Python Pandas Error tokenizing data and the different methods to solve it in different scenarios.
Different Errors while tokenizing data are,
Error tokenizing data. C error: Buffer overflow caught - possible malformed input fileParserError: Expected n fields in line x, saw mError tokenizing data. c error out of memory
Also learned the different engines available in the read_csv() method to parse the CSV file and the advantages and disadvantages of it.
You’ve also learned when to use the different methods appropriately.
If you have any questions, comment below.
You May Also Like
- How to write pandas dataframe to CSV
- How To Read An Excel File In Pandas – With Examples
- How To Read Excel With Multiple Sheets In Pandas?
- How To Solve Truth Value Of A Series Is Ambiguous. Use A.Empty, A.Bool(), A.Item(), A.Any() Or A.All()?
- How to solve xlrd.biffh.XLRDError: Excel xlsx file; not supported Error?
In this tutorial you’ll learn how to fix the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language.
The article consists of the following information:
Let’s get started.
Example Data & Software Libraries
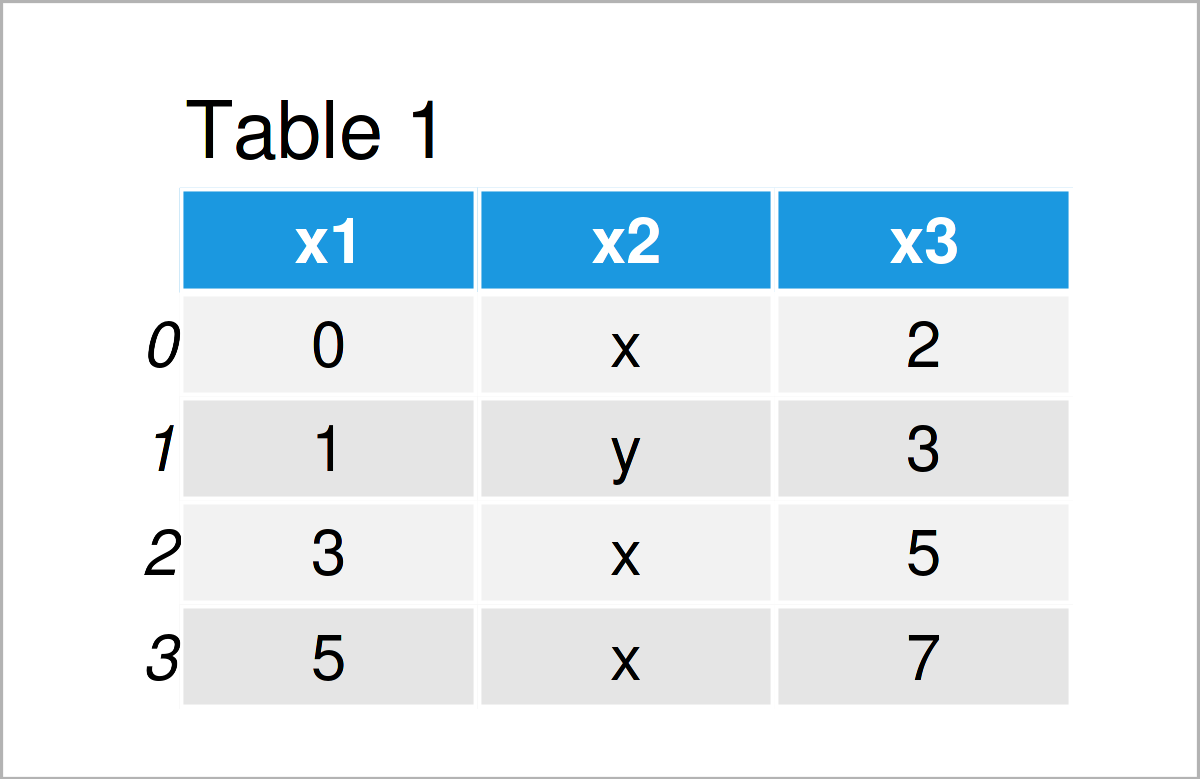
Consider the CSV file illustrated below as a basis for this tutorial:
You may already note that rows 4 and 6 contain one value too much. Those two rows contain four different values, but the other rows contain only three values.
Let’s assume that we want to read this CSV file as a pandas DataFrame into Python.
For this, we first have to import the pandas library:
import pandas as pd # Load pandas
Let’s move on to the examples!
Reproduce the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
In this section, I’ll show how to replicate the error message “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z”.
Let’s assume that we want to read our example CSV file using the default settings of the read_csv function. Then, we might try to import our data as shown below:
data_import = pd.read_csv('data.csv') # Try to import CSV file # ParserError: Error tokenizing data. C error: Expected 3 fields in line 4, saw 4
Unfortunately, the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” is returned after executing the Python syntax above.
The reason for this is that our CSV file contains too many values in some of the rows.
In the next section, I’ll show an easy solution for this problem. So keep on reading…
Debug the ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z
In this example, I’ll explain an easy fix for the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language.
We can ignore all lines in our CSV file that are formatted wrongly by specifying the error_bad_lines argument to False.
Have a look at the example code below:
data_import = pd.read_csv('data.csv', # Remove rows with errors error_bad_lines = False) print(data_import) # Print imported pandas DataFrame
As shown in Table 2, we have created a valid pandas DataFrame output using the previous code. As you can see, we have simply skipped the rows with too many values.
This is a simply trick that usually works. However, please note that this trick should be done with care, since the discussed error message typically points to more general issues with your data.
For that reason, it’s advisable to investigate why some of the rows are not formatted properly.
For this, I can also recommend this thread on Stack Overflow. It discusses how to identify wrong lines, and it also discusses other less common reasons for the error message “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z”.
Video & Further Resources
Have a look at the following video on my YouTube channel. In the video, I’m explaining the Python codes of this tutorial:
The YouTube video will be added soon.
Furthermore, you might read the other articles on this website. You can find some interesting tutorials below:
- Read CSV File as pandas DataFrame in Python
- Skip Rows but Keep Header when Reading CSV File
- Skip First Row when Reading pandas DataFrame from CSV File
- Specify Delimiter when Reading pandas DataFrame from CSV File
- Ignore Header when Reading CSV File as pandas DataFrame
- Check Data Type of Columns in pandas DataFrame in Python
- Change Data Type of pandas DataFrame Column in Python
- Basic Course for the pandas Library in Python
- Introduction to Python Programming
Summary: In this article, I have explained how to handle the “ParserError: Error tokenizing data. C error: Expected X fields in line Y, saw Z” in the Python programming language. If you have any further questions or comments, let me know in the comments. Furthermore, don’t forget to subscribe to my email newsletter to get updates on new articles.
Common reasons for ParserError: Error tokenizing data when initiating a Pandas DataFrame include:
-
Using the wrong delimiter
-
Number of fields in certain rows do not match with header
To resolve the error we can try the following:
-
Specifying the delimiter through
sepparameter inread_csv(~) -
Fixing the original source file
-
Skipping bad rows
Examples
Specifying sep
By default the read_csv(~) method assumes sep=",". Therefore when reading files that use a different delimiter, make sure to explicitly specify the delimiter to use.
Consider the following slash-delimited file called test.txt:
col1/col2/col3
1/A/4
2/B/5
3/C,D,E/6
To initialize a DataFrame using default sep=",":
import pandas as pd
df = pd.read_csv('test.txt')
ParserError: Error tokenizing data. C error: Expected 1 fields in line 4, saw 3
An error is raised as the first line in the file does not contain any commas, so read_csv(~) expects all lines in the file to only contain 1 field. However, line 4 has 3 fields as it contains two commas, which results in the ParserError.
To initialize the DataFrame by correctly specifying slash (/) as the delimiter:
df = pd.read_csv('test.txt', sep='/')
df
col1 col2 col3
0 1 A 4
1 2 B 5
2 3 C,D,E 6
We can now see the DataFrame is initialized as expected with each line containing the 3 fields which were separated by slashes (/) in the original file.
Fixing original source file
Consider the following comma-separated file called test.csv:
col1,col2,col3
1,A,4
2,B,5,
3,C,6
To initialize a DataFrame using the above file:
import pandas as pd
df = pd.read_csv('test.csv')
ParserError: Error tokenizing data. C error: Expected 3 fields in line 3, saw 4
Here there is an error on the 3rd line as 4 fields are observed instead of 3, caused by the additional comma at the end of the line.
To resolve this error, we can correct the original file by removing the extra comma at the end of line 3:
col1,col2,col3
1,A,4
2,B,5
3,C,6
To now initialize the DataFrame again using the corrected file:
df = pd.read_csv('test.csv')
df
col1 col2 col3
0 1 A 4
1 2 B 5
2 3 C 6
Skipping bad rows
Consider the following comma-separated file called test.csv:
col1,col2,col3
1,A,4
2,B,5,
3,C,6
To initialize a DataFrame using the above file:
import pandas as pd
df = pd.read_csv('test.csv')
ParserError: Error tokenizing data. C error: Expected 3 fields in line 3, saw 4
Here there is an error on the 3rd line as 4 fields are observed instead of 3, caused by the additional comma at the end of the line.
To skip bad rows pass on_bad_lines='skip' to read_csv(~):
df = pd.read_csv('test.csv', on_bad_lines='skip')
df
col1 col2 col3
0 1 A 4
1 3 C 6
Notice how the problematic third line in the original file has been skipped in the resulting DataFrame.
WARNING
This should be your last resort as valuable information could be contained within the problematic lines. Skipping these rows means you lose this information. As much as possible try to identify the root cause of the error and fix the underlying problem.
If engine='python' is used, curiously, it loads the DataFrame without any hiccups though. I used the following snippet to extract the first 3 lines in the file and 3 of the offending lines (got the line numbers from the error message).
from csv import reader
N = int(input('What line do you need? > '))
with open(filename) as f:
print(next((x for i, x in enumerate(reader(f)) if i == N), None))
Lines 1-3:
['08', '8', '7', '5', '0', '12', '54', '0', '11', '1', '58', '9', '68', '48.2', '0.756', '11.6', '17.5', '13.3', '4.3', '11.3', '32.2', '6.4', '4.1', '5.6', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '', '', '', '', '', '', '', '', '', '', '', '32']
['08', '8', '7', '5', '0', '15', '80', '0', '11', '1', '62', '9', '69', '77.8', '3.267', '11.2', '17.7', '14.8', '4.2', '15.2', '29.1', '18.4', '10.0', '18.1', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '', '', '', '', '', '', '', '', '', '', '', '32']
['08', '8', '7', '5', '0', '21', '52', '0', '11', '1', '61', '11', '51', '29.4', '0.076', '4.1', '13.8', '8.3', '21.5', '5.3', '3.1', '5.7', '3.0', '6.1', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '', '', '', '', '', '', '', '', '', '', '', '32']
Offending lines:
['09', '9', '15', '22', '46', '9', '51', '0', '11', '1', '57', '9', '70', '36.3', '0.242', '11.8', '16.2', '6.4', '4.1', '5.8', '31.3', '5.5', '3.9', '6.8', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '', '', '', '', '', '', '', '', '', '', '', '32']
['09', '9', '15', '22', '46', '25', '31', '0', '11', '1', '70', '9', '73', '67.8', '2.196', '10.4', '17.0', '13.4', '4.4', '12.2', '31.8', '15.6', '4.2', '16.2', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '', '', '', '', '', '', '', '', '', '', '', '32']
['09', '9', '15', '22', '46', '28', '41', '0', '11', '1', '70', '5', '22', '7.4', '0.003', '4.0', '13.1', '3.4', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '0.0', '', '', '', '', '', '', '', '', '', '', '', '32']
As you suggested, I will try to read the file, then modify the DataFrame (rename columns, delete unnecessary ones etc.) or simply use the python engine (long processing time).
Recently, I burned about 3 hours trying to load a large CSV file into Python Pandas using the read_csv function, only to consistently run into the following error:
ParserError Traceback (most recent call last) <ipython-input-6-b51ad8562823> in <module>() ... pandas_libsparsers.pyx in pandas._libs.parsers.TextReader.read (pandas_libsparsers.c:10862)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_low_memory (pandas_libsparsers.c:11138)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._read_rows (pandas_libsparsers.c:11884)() pandas_libsparsers.pyx in pandas._libs.parsers.TextReader._tokenize_rows (pandas_libsparsers.c:11755)() pandas_libsparsers.pyx in pandas._libs.parsers.raise_parser_error (pandas_libsparsers.c:28765)() Error tokenizing data. C error: EOF inside string starting at line XXXX
“Error tokenising data. C error: EOF inside string starting at line”.
There was an erroneous character about 5000 lines into the CSV file that prevented the Pandas CSV parser from reading the entire file. Excel had no problems opening the file, and no amount of saving/re-saving/changing encodings was working. Manually removing the offending line worked, but ultimately, another character 6000 lines further into the file caused the same issue.
The solution was to use the parameter engine=’python’ in the read_csv function call. The Pandas CSV parser can use two different “engines” to parse CSV file – Python or C (default).
pandas.read_csv(filepath_or_buffer, sep=', ', delimiter=None,
header='infer', names=None,
index_col=None, usecols=None, squeeze=False,
..., engine=None, ...)
engine : {‘c’, ‘python’}, optional
Parser engine to use. The C engine is faster while the python engine is currently more feature-complete.
The Python engine is described as “slower, but more feature complete” in the Pandas documentation. However, in this case, the python engine sorts the problem, without a massive time impact (overall file size was approx 200,000 rows).
UnicodeDecodeError
The other big problem with reading in CSV files that I come across is the error:
“UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position XX: invalid start byte”
Character encoding issues can be usually fixed by opening the CSV file in Sublime Text, and then “Save with encoding” choosing UTF-8. Then adding the encoding=’utf8′ to the pandas.read_csv command allows Pandas to open the CSV file without trouble. This error appears regularly for me with files saved in Excel.
encoding : str, default None